Gene Page: ESR1
Summary ?
| GeneID | 2099 |
| Symbol | ESR1 |
| Synonyms | ER|ESR|ESRA|ESTRR|Era|NR3A1 |
| Description | estrogen receptor 1 |
| Reference | MIM:133430|HGNC:HGNC:3467|Ensembl:ENSG00000091831|HPRD:00589|Vega:OTTHUMG00000016103 |
| Gene type | protein-coding |
| Map location | 6q25.1 |
| Pascal p-value | 0.85 |
| Fetal beta | -0.912 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 2.351 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.4727 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18007957 | 6 | 152011666 | ESR1 | 5.682E-4 | 0.422 | 0.049 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs443832 | chr16 | 83779643 | ESR1 | 2099 | 0.16 | trans |
Section II. Transcriptome annotation
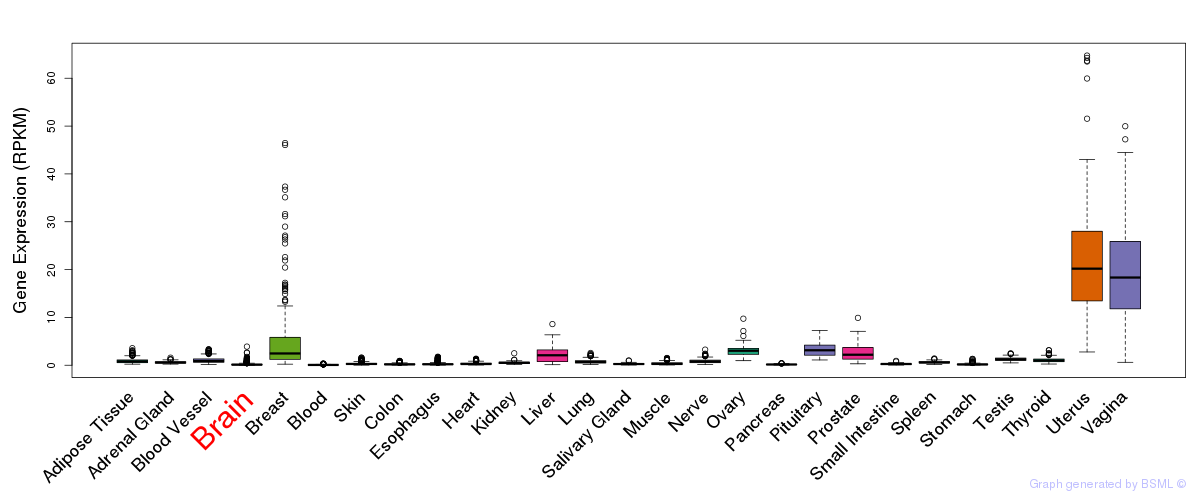
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CAMKK1 | 0.93 | 0.78 |
| SYT12 | 0.93 | 0.86 |
| EHD3 | 0.92 | 0.91 |
| MAST3 | 0.92 | 0.87 |
| SLC12A5 | 0.92 | 0.86 |
| CNTNAP1 | 0.91 | 0.87 |
| SIRPA | 0.91 | 0.90 |
| IQSEC1 | 0.91 | 0.85 |
| PHF15 | 0.91 | 0.82 |
| MAP1A | 0.90 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.54 | -0.65 |
| C9orf46 | -0.53 | -0.63 |
| GTF3C6 | -0.52 | -0.57 |
| EXOSC8 | -0.51 | -0.59 |
| FAM36A | -0.50 | -0.50 |
| DYNLT1 | -0.49 | -0.67 |
| AC006276.2 | -0.49 | -0.50 |
| RBMX2 | -0.48 | -0.62 |
| RPS8 | -0.48 | -0.65 |
| RPL36 | -0.48 | -0.64 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0003677 | DNA binding | IEA | - | |
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0003707 | steroid hormone receptor activity | IEA | - | |
| GO:0003707 | steroid hormone receptor activity | NAS | 11004670 | |
| GO:0030235 | nitric-oxide synthase regulator activity | NAS | 12389206 | |
| GO:0005496 | steroid binding | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0030284 | estrogen receptor activity | NAS | 3753802 | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0047485 | protein N-terminus binding | IPI | 12917342 | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | NAS | 3753802 | |
| GO:0030520 | estrogen receptor signaling pathway | NAS | 3753802 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0007165 | signal transduction | TAS | 10749889 |10970861 | |
| GO:0048386 | positive regulation of retinoic acid receptor signaling pathway | IDA | 15831516 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005634 | nucleus | NAS | - | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0005886 | plasma membrane | IDA | 18029348 | |
| GO:0016585 | chromatin remodeling complex | NAS | 12351687 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ER interacts with LOC441967. | BIND | 16009131 | |||
| AHR | - | aryl hydrocarbon receptor | - | HPRD,BioGRID | 10620335 |12612060 |
| AKAP13 | AKAP-Lbc | BRX | FLJ11952 | FLJ43341 | HA-3 | Ht31 | LBC | PROTO-LB | PROTO-LBC | c-lbc | A kinase (PRKA) anchor protein 13 | - | HPRD,BioGRID | 9627117 |
| ANP32A | C15orf1 | I1PP2A | LANP | MAPM | MGC119787 | MGC150373 | PHAP1 | PHAPI | PP32 | acidic (leucine-rich) nuclear phosphoprotein 32 family, member A | ER-alpha interacts with pp32. | BIND | 15308690 |
| AP1B1 | ADTB1 | AP105A | BAM22 | CLAPB2 | adaptor-related protein complex 1, beta 1 subunit | ER interacts with AP1B1. | BIND | 16009131 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 12389206 |
| ASCC2 | ASC1p100 | activating signal cointegrator 1 complex subunit 2 | ER interacts with ASC1p100. | BIND | 16009131 |
| AXIN2 | AXIL | DKFZp781B0869 | MGC10366 | MGC126582 | axin 2 | ER-alpha interacts with the Axin2 promoter. | BIND | 15304487 |
| BAG1 | RAP46 | BCL2-associated athanogene | - | HPRD,BioGRID | 8524784 |
| BAGE3 | - | B melanoma antigen family, member 3 | ER interacts with BAGE3. | BIND | 16009131 |
| BAZ1B | WBSCR10 | WBSCR9 | WSTF | bromodomain adjacent to zinc finger domain, 1B | Affinity Capture-Western | BioGRID | 12837248 |
| BCAS2 | DAM1 | breast carcinoma amplified sequence 2 | BCAS2 interacts with ER-alpha. This interaction was modeled on a demonstrated interaction between human BCAS2 and ER-alpha from an unspecified species. | BIND | 15694360 |
| BDNF | MGC34632 | brain-derived neurotrophic factor | - | HPRD | 11923430 |
| BLOC1S1 | BLOS1 | FLJ39337 | FLJ97089 | GCN5L1 | MGC87455 | MICoA | RT14 | biogenesis of lysosomal organelles complex-1, subunit 1 | ER interacts with MICoA | BIND | 12639951 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD | 10334989 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Affinity Capture-Western Co-localization Reconstituted Complex | BioGRID | 11244506 |11493692 |11782371 |12400015 |
| C21orf129 | FLJ32835 | PRED76 | chromosome 21 open reading frame 129 | ER interacts with C21orf129. | BIND | 16009131 |
| CALM1 | CALML2 | CAMI | DD132 | PHKD | calmodulin 1 (phosphorylase kinase, delta) | - | HPRD | 11981030 |
| CALM2 | CAMII | PHKD | PHKD2 | calmodulin 2 (phosphorylase kinase, delta) | - | HPRD | 11981030 |
| CALM3 | PHKD | PHKD3 | calmodulin 3 (phosphorylase kinase, delta) | - | HPRD | 11981030 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | - | HPRD,BioGRID | 11563984 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | - | HPRD,BioGRID | 9039267 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | ER-alpha interacts with cyclin D1 promoter. | BIND | 15808510 |
| CCNH | CAK | p34 | p37 | cyclin H | Affinity Capture-Western | BioGRID | 12527756 |
| CDC25B | - | cell division cycle 25 homolog B (S. pombe) | - | HPRD,BioGRID | 11689696 |
| CDK7 | CAK1 | CDKN7 | MO15 | STK1 | p39MO15 | cyclin-dependent kinase 7 | Biochemical Activity | BioGRID | 10949034 |
| CEBPA | C/EBP-alpha | CEBP | CCAAT/enhancer binding protein (C/EBP), alpha | Reconstituted Complex | BioGRID | 9817600 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | - | HPRD,BioGRID | 7651415 |
| CELSR1 | CDHF9 | DKFZp434P0729 | FMI2 | HFMI2 | ME2 | cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) | ER interacts with CELSR1. | BIND | 16009131 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | IKK-alpha interacts with ER-alpha. | BIND | 15808510 |
| COBRA1 | DKFZp586B0519 | KIAA1182 | NELF-B | NELFB | cofactor of BRCA1 | - | HPRD,BioGRID | 15342491 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | Affinity Capture-Western Reconstituted Complex | BioGRID | 11113179 |11782371 |
| CSNK1E | HCKIE | MGC10398 | casein kinase 1, epsilon | ER interacts with CSNK1E. | BIND | 16009131 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Affinity Capture-Western | BioGRID | 15304487 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | beta-catenin interacts with ER-alpha. | BIND | 15304487 |
| CYB5R3 | B5R | DIA1 | cytochrome b5 reductase 3 | ER interacts with CYB5R3. | BIND | 16009131 |
| DAP3 | DAP-3 | DKFZp686G12159 | MGC126058 | MGC126059 | MRP-S29 | MRPS29 | bMRP-10 | death associated protein 3 | Reconstituted Complex | BioGRID | 10903152 |
| DDX17 | DKFZp761H2016 | P72 | RH70 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | ESR1 (ER-alpha) interacts with an unspecified isoform of DDX17 (p72). This interaction was modeled on a demonstrated interaction between ESR1 from an unspecified species and DDX17 from an unspecified species. | BIND | 12738788 |
| DDX5 | DKFZp686J01190 | G17P1 | HLR1 | HUMP68 | p68 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | Affinity Capture-Western Two-hybrid | BioGRID | 10409727 |
| DDX5 | DKFZp686J01190 | G17P1 | HLR1 | HUMP68 | p68 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | ESR1 (ER-alpha) interacts with DDX5 (p68). This interaction was modeled on a demonstrated interaction between ESR1 from an unspecified species and DDX5 from an unspecified species. | BIND | 12738788 |
| DDX54 | DP97 | MGC2835 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 | - | HPRD,BioGRID | 12466272 |
| DNTTIP2 | ERBP | FCF2 | HSU15552 | LPTS-RP2 | MGC163494 | RP4-561L24.1 | TdIF2 | deoxynucleotidyltransferase, terminal, interacting protein 2 | - | HPRD,BioGRID | 15047147 |
| DSCAM | CHD2-42 | CHD2-52 | Down syndrome cell adhesion molecule | ER interacts with DSCAM-1 enhancer. | BIND | 16009131 |
| EBAG9 | EB9 | PDAF | RCAS1 | estrogen receptor binding site associated, antigen, 9 | ER-alpha interacts with EBAG9 promoter. | BIND | 15808510 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 11887937 |
| EIF3I | EIF3S2 | PRO2242 | TRIP-1 | TRIP1 | eIF3-beta | eIF3-p36 | eukaryotic translation initiation factor 3, subunit I | ER alpha interacts with TRIP1. This interaction was modeled on a demonstrated interaction between TRIP1 from an unspecified source and human ER alpha. | BIND | 10706629 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | - | HPRD,BioGRID | 11782371 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | EP300 (p300) acetylates ESR1 (ER-alpha). This interaction was modeled on a demonstrated interaction between EP300 from an unspecified species and ESR1 from an unspecified species. | BIND | 14761960 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | Two-hybrid | BioGRID | 12198596 |
| ESR2 | ER-BETA | ESR-BETA | ESRB | ESTRB | Erb | NR3A2 | estrogen receptor 2 (ER beta) | ER alpha interacts with ER beta to form heterodimer. This interaction was modeled on a demonstrated interaction between human ER alpha and mouse ER beta. | BIND | 10706629 |
| ESR2 | ER-BETA | ESR-BETA | ESRB | ESTRB | Erb | NR3A2 | estrogen receptor 2 (ER beta) | - | HPRD,BioGRID | 9473491 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | ER-alpha interacts with FHL2. | BIND | 15666801 |
| FKBP4 | FKBP52 | FKBP59 | HBI | Hsp56 | PPIase | p52 | FK506 binding protein 4, 59kDa | Reconstituted Complex | BioGRID | 9222609 |
| FKBP5 | FKBP51 | FKBP54 | MGC111006 | P54 | PPIase | Ptg-10 | FK506 binding protein 5 | Reconstituted Complex | BioGRID | 9222609 |
| FOXO1 | FKH1 | FKHR | FOXO1A | forkhead box O1 | - | HPRD,BioGRID | 11435445 |
| FOXO4 | AFX | AFX1 | MGC120490 | MLLT7 | forkhead box O4 | - | HPRD,BioGRID | 11435445 |
| GADD45A | DDIT1 | GADD45 | growth arrest and DNA-damage-inducible, alpha | Affinity Capture-Western | BioGRID | 10872826 |
| GADD45B | DKFZp566B133 | GADD45BETA | MYD118 | growth arrest and DNA-damage-inducible, beta | - | HPRD | 10872826 |
| GADD45G | CR6 | DDIT2 | GADD45gamma | GRP17 | growth arrest and DNA-damage-inducible, gamma | - | HPRD,BioGRID | 10872826 |
| GTF2H1 | BTF2 | TFB1 | TFIIH | general transcription factor IIH, polypeptide 1, 62kDa | - | HPRD,BioGRID | 10949034 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | ER-alpha interacts with HDAC1. | BIND | 14506733 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD,BioGRID | 11911945 |
| HSPA4 | APG-2 | HS24/P52 | MGC131852 | RY | hsp70 | hsp70RY | heat shock 70kDa protein 4 | Reconstituted Complex | BioGRID | 9222609 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | Affinity Capture-Western | BioGRID | 16113100 |
| ISL1 | Isl-1 | ISL LIM homeobox 1 | - | HPRD,BioGRID | 11043578 |
| JARID1A | KDM5A | RBBP2 | RBP2 | jumonji, AT rich interactive domain 1A | Affinity Capture-Western Reconstituted Complex | BioGRID | 11358960 |
| JARID1A | KDM5A | RBBP2 | RBP2 | jumonji, AT rich interactive domain 1A | Rbp2 interacts with ER. | BIND | 11358960 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | ER-alpha interacts with c-Jun. This interaction was modeled on a demonstrated interaction between human ER-alpha, and mouse c-Jun. | BIND | 11477071 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 11477071 |
| JUNB | AP-1 | jun B proto-oncogene | ER-alpha interacts with JunB. This interaction was modeled on a demonstrated interaction between human ER-alpha and mouse JunB. | BIND | 11477071 |
| LOH3CR2A | ERR-10 | ERR10 | NAG-7 | NAG7 | loss of heterozygosity, 3, chromosomal region 2, gene A | - | HPRD | 15474036 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | - | HPRD,BioGRID | 12093745 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Reconstituted Complex | BioGRID | 10766163 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | ESR1 (ER-alpha) interacts with PPARBP (TRAP220). This interaction was modeled on a demonstrated interaction between ESR1 from an unspecified species and PPARBP from an unspecified species. | BIND | 12738788 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | Affinity Capture-Western | BioGRID | 12837248 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | - | HPRD | 9653119 |
| MED24 | ARC100 | CRSP100 | CRSP4 | DRIP100 | KIAA0130 | MGC8748 | THRAP4 | TRAP100 | mediator complex subunit 24 | - | HPRD | 9653119 |
| MED24 | ARC100 | CRSP100 | CRSP4 | DRIP100 | KIAA0130 | MGC8748 | THRAP4 | TRAP100 | mediator complex subunit 24 | Affinity Capture-MS | BioGRID | 12837248 |
| MGMT | - | O-6-methylguanine-DNA methyltransferase | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 11564893 |
| MKNK2 | GPRK7 | MNK2 | MAP kinase interacting serine/threonine kinase 2 | - | HPRD,BioGRID | 11013076 |
| MMS19 | FLJ34167 | FLJ95146 | MET18 | MGC99604 | MMS19L | hMMS19 | MMS19 nucleotide excision repair homolog (S. cerevisiae) | - | HPRD | 11279242 |
| MNAT1 | MAT1 | RNF66 | TFB3 | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | - | HPRD,BioGRID | 12527756 |
| MPG | AAG | APNG | CRA36.1 | MDG | Mid1 | PIG11 | PIG16 | anpg | N-methylpurine-DNA glycosylase | ESR1 (ER-alpha) interacts with MPG. | BIND | 14761960 |
| MTA1 | - | metastasis associated 1 | - | HPRD,BioGRID | 12167865 |
| MVP | LRP | VAULT1 | major vault protein | - | HPRD,BioGRID | 9628887 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | ER-alpha interacts with c-myc promoter. | BIND | 15808510 |
| NCAM2 | MGC51008 | NCAM21 | neural cell adhesion molecule 2 | ER interacts with NCAM2. | BIND | 16009131 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | ER-alpha interacts with SRC-1 | BIND | 10938099 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD | 9427757 |11165056 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | Reconstituted Complex | BioGRID | 9427757 |11003650 |11113179 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | ER alpha interacts with TIF2. This interaction was modeled on a demonstrated interaction between TIF2 from an unspecified source and human ER alpha. | BIND | 10706629 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | - | HPRD,BioGRID | 11937504 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | - | HPRD,BioGRID | 11050174 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | SRC-3 interacts with ER. This interaction was modelled on a demonstrated interaction between human SRC-3 and ER from an unspecified species. | BIND | 15383283 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | TRAM-1 interacts with ER. | BIND | 9346901 |
| NCOA4 | ARA70 | DKFZp762E1112 | ELE1 | PTC3 | RFG | nuclear receptor coactivator 4 | Reconstituted Complex | BioGRID | 9892017 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | Reconstituted Complex Two-hybrid | BioGRID | 10567404 |11158331 |11773444 |
| NCOA7 | ERAP140 | ESNA1 | FLJ45605 | MGC88425 | Nbla00052 | Nbla10993 | dJ187J11.3 | nuclear receptor coactivator 7 | - | HPRD,BioGRID | 11971969 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | - | HPRD,BioGRID | 12145334 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | ESR1 (ER-alpha) interacts with NCOR1 (N-CoR). This interaction was modeled on a demonstrated interaction between ESR1 from an unspecified species and human NCOR1. | BIND | 12738788 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | ESR1 (ER-alpha) interacts with NCOR2 (SMRT). This interaction was modeled on a demonstrated interaction between ESR1 from an unspecified species and NCOR2 from an unspecified species. | BIND | 12738788 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | - | HPRD | 9702189 |
| NR0B2 | FLJ17090 | SHP | SHP1 | nuclear receptor subfamily 0, group B, member 2 | - | HPRD,BioGRID | 9773978 |11861507 |
| NR2C1 | TR2 | nuclear receptor subfamily 2, group C, member 1 | - | HPRD,BioGRID | 12093804 |
| NR2C2 | TAK1 | TR2R1 | TR4 | hTAK1 | nuclear receptor subfamily 2, group C, member 2 | - | HPRD,BioGRID | 11844790 |
| NR2F1 | COUP-TFI | EAR-3 | EAR3 | ERBAL3 | NR2F2 | SVP44 | TCFCOUP1 | TFCOUP1 | nuclear receptor subfamily 2, group F, member 1 | - | HPRD,BioGRID | 12093745 |
| NR2F6 | EAR-2 | EAR2 | ERBAL2 | nuclear receptor subfamily 2, group F, member 6 | - | HPRD,BioGRID | 10713182 |
| NRIP1 | FLJ77253 | RIP140 | nuclear receptor interacting protein 1 | ESR1 (ER-alpha) interacts with NRIP1 (RIP140). This interaction was modelled on a demonstrated interaction between human ESR1 and mouse NRIP1. | BIND | 9774463 |
| NRIP1 | FLJ77253 | RIP140 | nuclear receptor interacting protein 1 | ER interacts with NRIP-1 enhancer. | BIND | 16009131 |
| NRIP1 | FLJ77253 | RIP140 | nuclear receptor interacting protein 1 | - | HPRD,BioGRID | 7641693 |
| NSD1 | ARA267 | DKFZp666C163 | FLJ10684 | FLJ22263 | FLJ44628 | KMT3B | SOTOS | STO | nuclear receptor binding SET domain protein 1 | - | HPRD | 9628876 |
| PAK6 | PAK5 | p21 protein (Cdc42/Rac)-activated kinase 6 | - | HPRD,BioGRID | 11773441 |
| PELP1 | HMX3 | MNAR | P160 | proline, glutamate and leucine rich protein 1 | - | HPRD,BioGRID | 11481323 |
| PGR | NR3C3 | PR | progesterone receptor | ER-alpha interacts with the PgRA promoter. | BIND | 15019994 |
| PGR | NR3C3 | PR | progesterone receptor | ER-alpha interacts with the PgRB promoter. | BIND | 15019994 |
| PHB2 | BAP | BCAP37 | Bap37 | MGC117268 | PNAS-141 | REA | p22 | prohibitin 2 | ER-alpha interacts with REA | BIND | 10938099 |
| PHB2 | BAP | BCAP37 | Bap37 | MGC117268 | PNAS-141 | REA | p22 | prohibitin 2 | Two-hybrid | BioGRID | 12943695 |
| PIAS1 | DDXBP1 | GBP | GU/RH-II | MGC141878 | MGC141879 | ZMIZ3 | protein inhibitor of activated STAT, 1 | ER-alpha interacts with PIAS1. | BIND | 15666801 |
| PIAS1 | DDXBP1 | GBP | GU/RH-II | MGC141878 | MGC141879 | ZMIZ3 | protein inhibitor of activated STAT, 1 | - | HPRD | 15666801 |
| PIAS2 | MGC102682 | MIZ1 | PIASX | PIASX-ALPHA | PIASX-BETA | SIZ2 | ZMIZ4 | miz | protein inhibitor of activated STAT, 2 | Reconstituted Complex | BioGRID | 11117529 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD,BioGRID | 11689445 |
| PNRC2 | FLJ20312 | MGC99541 | proline-rich nuclear receptor coactivator 2 | - | HPRD | 11574675 |
| POU2F1 | OCT1 | OTF1 | POU class 2 homeobox 1 | Reconstituted Complex | BioGRID | 10480874 |
| POU2F2 | OCT2 | OTF2 | Oct-2 | POU class 2 homeobox 2 | Reconstituted Complex | BioGRID | 10480874 |
| POU4F1 | BRN3A | FLJ13449 | Oct-T1 | RDC-1 | POU class 4 homeobox 1 | Reconstituted Complex Two-hybrid | BioGRID | 9448000 |
| POU4F2 | BRN3.2 | BRN3B | Brn-3b | POU class 4 homeobox 2 | - | HPRD,BioGRID | 9448000 |
| PPARGC1A | LEM6 | PGC-1(alpha) | PGC-1v | PGC1 | PGC1A | PPARGC1 | peroxisome proliferator-activated receptor gamma, coactivator 1 alpha | - | HPRD,BioGRID | 10748020 |
| PPARGC1B | ERRL1 | PERC | PGC-1(beta) | PGC1B | peroxisome proliferator-activated receptor gamma, coactivator 1 beta | - | HPRD,BioGRID | 11854298 |
| PPID | CYP-40 | CYPD | MGC33096 | peptidylprolyl isomerase D | Reconstituted Complex | BioGRID | 9222609 |
| PRDM2 | HUMHOXY1 | KMT8 | MTB-ZF | RIZ | RIZ1 | RIZ2 | PR domain containing 2, with ZNF domain | - | HPRD,BioGRID | 10706618 |
| PRMT1 | ANM1 | HCP1 | HRMT1L2 | IR1B4 | protein arginine methyltransferase 1 | Reconstituted Complex | BioGRID | 11050077 |
| PRMT2 | HRMT1L1 | MGC111373 | protein arginine methyltransferase 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12039952 |
| PRMT2 | HRMT1L1 | MGC111373 | protein arginine methyltransferase 2 | PRMT2 interacts with ER-alpha. | BIND | 12039952 |
| PRPF6 | ANT-1 | C20orf14 | TOM | U5-102K | hPrp6 | PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) | - | HPRD | 12039962 |
| PTEN | 10q23del | BZS | MGC11227 | MHAM | MMAC1 | PTEN1 | TEP1 | phosphatase and tensin homolog | Reconstituted Complex | BioGRID | 15205473 |
| PTGES3 | P23 | TEBP | cPGES | prostaglandin E synthase 3 (cytosolic) | Reconstituted Complex | BioGRID | 9222609 |
| PTGES3 | P23 | TEBP | cPGES | prostaglandin E synthase 3 (cytosolic) | - | HPRD | 10207098 |
| PTMA | MGC104802 | TMSA | prothymosin, alpha | Reconstituted Complex | BioGRID | 12943695 |
| RBM23 | CAPERbeta | FLJ10482 | MGC4458 | PP239 | RNPC4 | RNA binding motif protein 23 | RBM23 (CAPER-beta) interacts with ESR1(ER-alpha) | BIND | 15694343 |
| RBM39 | CAPER | CAPERalpha | CC1.3 | DKFZp781C0423 | FLJ44170 | HCC1 | RNPC2 | fSAP59 | RNA binding motif protein 39 | - | HPRD,BioGRID | 11704680 |
| RBM9 | FOX2 | Fox-2 | HNRBP2 | HRNBP2 | RTA | dJ106I20.3 | fxh | RNA binding motif protein 9 | - | HPRD,BioGRID | 11875103 |
| REXO4 | REX4 | XPMC2 | XPMC2H | REX4, RNA exonuclease 4 homolog (S. cerevisiae) | - | HPRD,BioGRID | 10908561 |
| RFX4 | NYD-SP10 | regulatory factor X, 4 (influences HLA class II expression) | - | HPRD | 1603086 |10706293 |
| RNF14 | ARA54 | FLJ26004 | HFB30 | HRIHFB2038 | TRIAD2 | ring finger protein 14 | - | HPRD | 10085091 |
| RNF4 | RES4-26 | SNURF | ring finger protein 4 | Two-hybrid | BioGRID | 9710597 |
| SAFB | DKFZp779C1727 | HAP | HET | SAFB1 | scaffold attachment factor B | - | HPRD,BioGRID | 10707955 |
| SAFB2 | KIAA0138 | scaffold attachment factor B2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12660241 |
| SFI1 | MGC131712 | MGC150663 | MGC156283 | MGC57874 | PISD | RP5-858B16.1 | hSfi1p | Sfi1 homolog, spindle assembly associated (yeast) | ER interacts with SFI1. | BIND | 16009131 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 11773443 |
| SLC37A1 | FLJ22340 | G3PP | solute carrier family 37 (glycerol-3-phosphate transporter), member 1 | ER interacts with SLC37A1. | BIND | 16009131 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD,BioGRID | 11555647 |
| SMARCA2 | BAF190 | BRM | FLJ36757 | MGC74511 | SNF2 | SNF2L2 | SNF2LA | SWI2 | Sth1p | hBRM | hSNF2a | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | - | HPRD,BioGRID | 9099865 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | - | HPRD,BioGRID | 9099865 |
| SMARCD1 | BAF60A | CRACD1 | Rsc6p | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 | Reconstituted Complex | BioGRID | 12917342 |
| SMARCE1 | BAF57 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 | - | HPRD,BioGRID | 12145209 |
| SNAP29 | CEDNIK | FLJ21051 | SNAP-29 | synaptosomal-associated protein, 29kDa | ER interacts with SNAP29. | BIND | 16009131 |
| SNURF | - | SNRPN upstream reading frame | Two-hybrid | BioGRID | 11696545 |
| SOD1 | ALS | ALS1 | IPOA | SOD | homodimer | superoxide dismutase 1, soluble | ER interacts with SOD-1 enhancer. | BIND | 16009131 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Affinity Capture-Western Reconstituted Complex | BioGRID | 10454579 |11032808 |11564893 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | Affinity Capture-Western | BioGRID | 11429412 |
| STAT5A | MGF | STAT5 | signal transducer and activator of transcription 5A | ER-alpha interacts with STAT5a. | BIND | 15304355 |
| STAT5A | MGF | STAT5 | signal transducer and activator of transcription 5A | Reconstituted Complex | BioGRID | 11682624 |
| STRN | MGC125642 | SG2NA | striatin, calmodulin binding protein | - | HPRD | 15569929 |
| SVIL | DKFZp686A17191 | supervillin | Two-hybrid | BioGRID | 11792840 |
| TADA3L | ADA3 | FLJ20221 | FLJ21329 | hADA3 | transcriptional adaptor 3 (NGG1 homolog, yeast)-like | - | HPRD,BioGRID | 12034840 |
| TAF10 | TAF2A | TAF2H | TAFII30 | TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa | - | HPRD | 7923369 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | - | HPRD,BioGRID | 11595744 |
| TDG | - | thymine-DNA glycosylase | - | HPRD,BioGRID | 12874288 |
| TFF1 | BCEI | D21S21 | HP1.A | HPS2 | pNR-2 | pS2 | trefoil factor 1 | ER-alpha interacts with pS2. | BIND | 10490106 |
| TFF1 | BCEI | D21S21 | HP1.A | HPS2 | pNR-2 | pS2 | trefoil factor 1 | ER interacts with TFF-1 promoter. | BIND | 16009131 |
| TFF1 | BCEI | D21S21 | HP1.A | HPS2 | pNR-2 | pS2 | trefoil factor 1 | ER interacts with TFF-1 enhancer. | BIND | 16009131 |
| TFF1 | BCEI | D21S21 | HP1.A | HPS2 | pNR-2 | pS2 | trefoil factor 1 | ER-alpha interacts with the pS2 promoter. | BIND | 15304487 |
| THOC5 | C22orf19 | Fmip | PK1.3 | fSAP79 | THO complex 5 | ER interacts with C22orf19. | BIND | 16009131 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 10766163 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | P53 interacts with ER-alpha. | BIND | 10766163 |
| TRIM24 | PTC6 | RNF82 | TF1A | TIF1 | TIF1A | TIF1ALPHA | hTIF1 | tripartite motif-containing 24 | Reconstituted Complex | BioGRID | 9115274 |10598587 |
| TRIM24 | PTC6 | RNF82 | TF1A | TIF1 | TIF1A | TIF1ALPHA | hTIF1 | tripartite motif-containing 24 | - | HPRD | 9115274 |9632676 |10598587 |
| TRIM24 | PTC6 | RNF82 | TF1A | TIF1 | TIF1A | TIF1ALPHA | hTIF1 | tripartite motif-containing 24 | ESR1 (ER-alpha) interacts with TIF1. This interaction was modeled on a demonstrated interaction between human ESR1 and mouse TIF1. | BIND | 9774463 |
| TRIP4 | HsT17391 | thyroid hormone receptor interactor 4 | - | HPRD,BioGRID | 10454579 |
| TRRAP | FLJ10671 | PAF350/400 | PAF400 | STAF40 | TR-AP | Tra1 | transformation/transcription domain-associated protein | ESR1 (ER-alpha) interacts with TRRAP. This interaction was modeled on a demonstrated interaction between ESR1 from an unspecified species and TRRAP from an unspecified species. | BIND | 12738788 |
| TRRAP | FLJ10671 | PAF350/400 | PAF400 | STAF40 | TR-AP | Tra1 | transformation/transcription domain-associated protein | Affinity Capture-MS | BioGRID | 12837248 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | ER-alpha interacts with an unspecified isoform of Ubc9. | BIND | 15666801 |
| XBP1 | TREB5 | XBP2 | X-box binding protein 1 | - | HPRD,BioGRID | 12954762 |
| XBP1 | TREB5 | XBP2 | X-box binding protein 1 | ER interacts with XBP-1 enhancer. | BIND | 16009131 |
| YWHAH | YWHA1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | - | HPRD | 11266503 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | Reconstituted Complex | BioGRID | 11266503 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | - | HPRD,BioGRID | 14521715 |
| ZNF398 | KIAA1339 | P51 | P71 | ZER6 | zinc finger protein 398 | - | HPRD,BioGRID | 11779858 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA CARM ER PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MTA3 PATHWAY | 19 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HER2 PATHWAY | 22 | 17 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSII PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID ER NONGENOMIC PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID FOXM1 PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| PID ERA GENOMIC PATHWAY | 65 | 37 | All SZGR 2.0 genes in this pathway |
| PID P38 ALPHA BETA DOWNSTREAM PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PID HNF3A PATHWAY | 44 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR SIGNALING BY ERBB4 | 38 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | 49 | 36 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 UP | 112 | 72 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED UP | 78 | 48 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION UP | 119 | 66 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA DN | 146 | 94 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG DN | 59 | 40 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A DN | 90 | 61 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION 12HR | 43 | 35 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK TARGETS | 18 | 15 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| JAERVINEN AMPLIFIED IN LARYNGEAL CANCER | 40 | 24 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL DN | 114 | 58 | All SZGR 2.0 genes in this pathway |
| DAWSON METHYLATED IN LYMPHOMA TCL1 | 59 | 45 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 UP | 36 | 22 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| OHM METHYLATED IN ADULT CANCERS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER DN | 36 | 24 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION UP | 48 | 26 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 6HR | 59 | 38 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS | 108 | 71 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 3HR | 74 | 47 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER UP | 27 | 20 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| HOQUE METHYLATED IN CANCER | 56 | 45 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN DN | 85 | 58 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE UP | 97 | 61 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL A UP | 84 | 52 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| BUCKANOVICH T LYMPHOCYTE HOMING ON TUMOR UP | 24 | 11 | All SZGR 2.0 genes in this pathway |
| MATZUK PREOVULATORY FOLLICLE | 10 | 8 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 UP | 167 | 99 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN | 134 | 83 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL DN | 76 | 51 | All SZGR 2.0 genes in this pathway |
| LI PROSTATE CANCER EPIGENETIC | 30 | 22 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL UP | 193 | 95 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 2 | 86 | 50 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 3 | 15 | 7 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR DN | 88 | 59 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS DN | 116 | 74 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 DN | 88 | 61 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 DN | 82 | 51 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE UP | 116 | 65 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER CLUSTER 6 | 16 | 13 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-130/301 | 3766 | 3772 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-148/152 | 3767 | 3773 | m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-18 | 1938 | 1945 | 1A,m8 | hsa-miR-18a | UAAGGUGCAUCUAGUGCAGAUA |
| hsa-miR-18b | UAAGGUGCAUCUAGUGCAGUUA | ||||
| miR-181 | 1998 | 2005 | 1A,m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-188 | 196 | 202 | m8 | hsa-miR-188 | CAUCCCUUGCAUGGUGGAGGGU |
| miR-19 | 4216 | 4222 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA | ||||
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-203.1 | 2165 | 2172 | 1A,m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-204/211 | 195 | 201 | m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-219 | 2244 | 2250 | m8 | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-22 | 2292 | 2299 | 1A,m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-221/222 | 2253 | 2260 | 1A,m8 | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-26 | 3982 | 3988 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-330 | 4182 | 4188 | m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-361 | 3233 | 3239 | 1A | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
| miR-496 | 3841 | 3847 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-9 | 3086 | 3092 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 379 | 385 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.