Gene Page: FGF1
Summary ?
| GeneID | 2246 |
| Symbol | FGF1 |
| Synonyms | AFGF|ECGF|ECGF-beta|ECGFA|ECGFB|FGF-1|FGF-alpha|FGFA|GLIO703|HBGF-1|HBGF1 |
| Description | fibroblast growth factor 1 |
| Reference | MIM:131220|HGNC:HGNC:3665|Ensembl:ENSG00000113578|HPRD:00567|Vega:OTTHUMG00000059703 |
| Gene type | protein-coding |
| Map location | 5q31 |
| Pascal p-value | 0.216 |
| Fetal beta | -4.468 |
| Support | NEUROTROPHIN SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00459 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01718 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
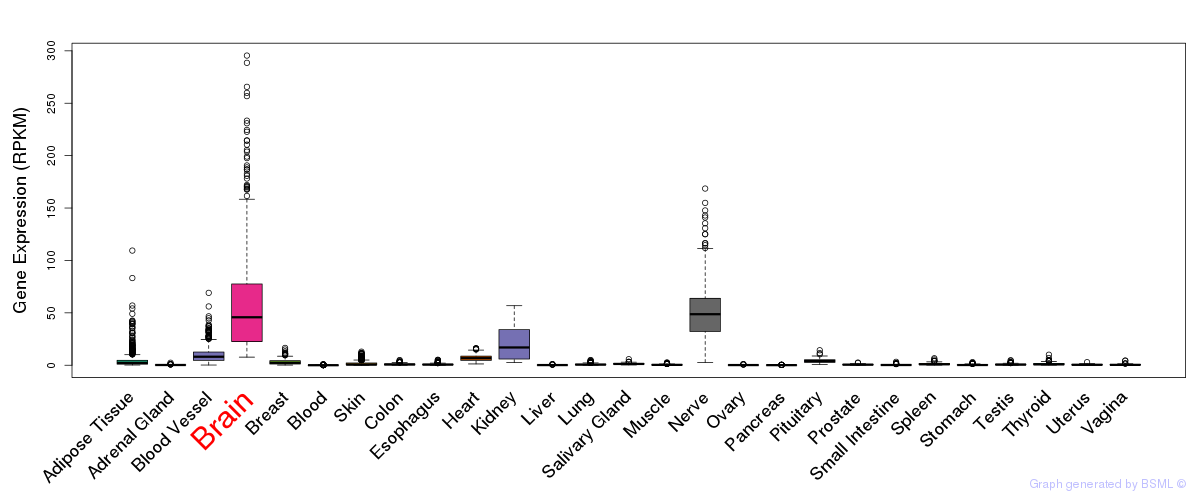
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LRIG1 | 0.81 | 0.80 |
| ACSS1 | 0.79 | 0.91 |
| INPPL1 | 0.77 | 0.78 |
| ACACB | 0.74 | 0.76 |
| STON2 | 0.74 | 0.76 |
| AXL | 0.74 | 0.87 |
| TRIP6 | 0.73 | 0.81 |
| NOTCH3 | 0.73 | 0.63 |
| RFX4 | 0.73 | 0.80 |
| PBXIP1 | 0.72 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ACTR10 | -0.42 | -0.49 |
| C20orf7 | -0.42 | -0.56 |
| RALYL | -0.42 | -0.54 |
| VPS29 | -0.42 | -0.52 |
| NHEDC2 | -0.42 | -0.55 |
| MED19 | -0.42 | -0.62 |
| TAF9 | -0.42 | -0.57 |
| HMGCLL1 | -0.42 | -0.56 |
| FARSB | -0.41 | -0.57 |
| ANKRD46 | -0.41 | -0.56 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 10618369 |14732692 | |
| GO:0008201 | heparin binding | IEA | - | |
| GO:0008083 | growth factor activity | IEA | - | |
| GO:0008083 | growth factor activity | TAS | 1693186 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001525 | angiogenesis | IEA | - | |
| GO:0001759 | induction of an organ | IEA | - | |
| GO:0007165 | signal transduction | NAS | 3778488 | |
| GO:0008283 | cell proliferation | IEA | - | |
| GO:0008543 | fibroblast growth factor receptor signaling pathway | EXP | 17133345 | |
| GO:0009653 | anatomical structure morphogenesis | TAS | 1693186 | |
| GO:0007275 | multicellular organismal development | TAS | 10903182 | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0030324 | lung development | IEA | - | |
| GO:0050679 | positive regulation of epithelial cell proliferation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | EXP | 1697263 |8622701 |9139660 |11294897 |16597617 | |
| GO:0005578 | proteinaceous extracellular matrix | IEA | - | |
| GO:0005615 | extracellular space | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | - | HPRD,BioGRID | 12145206 |
| CSNK2A2 | CK2A2 | CSNK2A1 | FLJ43934 | casein kinase 2, alpha prime polypeptide | - | HPRD,BioGRID | 12145206 |
| CSNK2B | CK2B | CK2N | CSK2B | G5A | MGC138222 | MGC138224 | casein kinase 2, beta polypeptide | - | HPRD,BioGRID | 12145206 |
| FGF1 | AFGF | ECGF | ECGF-beta | ECGFA | ECGFB | FGF-alpha | FGFA | GLIO703 | HBGF1 | fibroblast growth factor 1 (acidic) | - | HPRD,BioGRID | 9655399|11069186 |
| FGFBP1 | FGFBP | HBP17 | fibroblast growth factor binding protein 1 | - | HPRD,BioGRID | 11509569 |
| FGFR1 | BFGFR | CD331 | CEK | FGFBR | FLG | FLJ99988 | FLT2 | HBGFR | KAL2 | N-SAM | fibroblast growth factor receptor 1 | Co-crystal Structure Reconstituted Complex | BioGRID | 8576175 |11030354 |
| FGFR1 | BFGFR | CD331 | CEK | FGFBR | FLG | FLJ99988 | FLT2 | HBGFR | KAL2 | N-SAM | fibroblast growth factor receptor 1 | - | HPRD | 10336501 |11032250 |
| FGFR2 | BEK | BFR-1 | CD332 | CEK3 | CFD1 | ECT1 | FLJ98662 | JWS | K-SAM | KGFR | TK14 | TK25 | fibroblast growth factor receptor 2 | - | HPRD | 10618369|11069186 |
| FGFR2 | BEK | BFR-1 | CD332 | CEK3 | CFD1 | ECT1 | FLJ98662 | JWS | K-SAM | KGFR | TK14 | TK25 | fibroblast growth factor receptor 2 | Co-crystal Structure Reconstituted Complex | BioGRID | 8576175 |10618369 |11069186 |
| FGFR3 | ACH | CD333 | CEK2 | HSFGFR3EX | JTK4 | fibroblast growth factor receptor 3 | - | HPRD,BioGRID | 10574949 |
| FGFR4 | CD334 | JTK2 | MGC20292 | TKF | fibroblast growth factor receptor 4 | - | HPRD,BioGRID | 10336501 |10736564 |
| FIBP | FGFIBP | FIBP-1 | fibroblast growth factor (acidic) intracellular binding protein | - | HPRD,BioGRID | 9806903 |
| HSPA9 | CSA | GRP75 | HSPA9B | MGC4500 | MOT | MOT2 | MTHSP75 | PBP74 | mot-2 | heat shock 70kDa protein 9 (mortalin) | - | HPRD,BioGRID | 10510314 |
| NRP1 | BDCA4 | CD304 | DKFZp686A03134 | DKFZp781F1414 | NP1 | NRP | VEGF165R | neuropilin 1 | NRP1 (Npn-1) interacts with FGF1. This interaction was modeled on a demonstrated interaction between rat NRP1 and human FGF1. | BIND | 15695515 |
| S100A13 | - | S100 calcium binding protein A13 | - | HPRD,BioGRID | 9712836 |11432880|11432880 |
| SYT1 | DKFZp781D2042 | P65 | SVP65 | SYT | synaptotagmin I | - | HPRD,BioGRID | 11432880 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOMA | 71 | 57 | All SZGR 2.0 genes in this pathway |
| PID FGF PATHWAY | 55 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | 37 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | 87 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | 16 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR1 MUTANTS | 30 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR3 MUTANTS | 11 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR MUTANTS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME FRS2 MEDIATED CASCADE | 36 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME PI 3K CASCADE | 56 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | 100 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPASE C MEDIATED CASCADE | 54 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME SHC MEDIATED CASCADE | 28 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY INSULIN RECEPTOR | 108 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME FGFR LIGAND BINDING AND ACTIVATION | 22 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | 12 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | 14 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K CASCADE | 71 | 51 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR UP | 162 | 116 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS UP | 82 | 58 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| KHETCHOUMIAN TRIM24 TARGETS DN | 8 | 7 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| HE PTEN TARGETS UP | 16 | 15 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS DN | 108 | 84 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| GRANDVAUX IRF3 TARGETS DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| MCDOWELL ACUTE LUNG INJURY DN | 48 | 33 | All SZGR 2.0 genes in this pathway |
| WENG POR TARGETS LIVER DN | 20 | 11 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS FIBROBLAST UP | 84 | 60 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K4ME3 | 174 | 100 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES LCP WITH H3K4ME3 | 142 | 80 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC LCP WITH H3K4ME3 | 58 | 34 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 14 | 143 | 86 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| KANG AR TARGETS DN | 19 | 12 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-143 | 1432 | 1438 | m8 | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.