| PID REG GR PATHWAY
| 82 | 60 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP
| 294 | 178 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP
| 579 | 346 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP
| 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN UP
| 184 | 125 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN
| 367 | 220 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 6HR UP
| 85 | 54 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS DN
| 139 | 76 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN
| 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN
| 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN
| 537 | 339 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN
| 485 | 334 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS UP
| 77 | 62 | All SZGR 2.0 genes in this pathway |
| HWANG PROSTATE CANCER MARKERS
| 28 | 19 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL
| 326 | 213 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS
| 58 | 45 | All SZGR 2.0 genes in this pathway |
| LIANG HEMATOPOIESIS STEM CELL NUMBER LARGE VS TINY DN
| 45 | 24 | All SZGR 2.0 genes in this pathway |
| FALVELLA SMOKERS WITH LUNG CANCER
| 80 | 52 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN
| 1024 | 594 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR UP
| 116 | 79 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP
| 487 | 286 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN
| 411 | 249 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1
| 379 | 235 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP
| 256 | 159 | All SZGR 2.0 genes in this pathway |
| NELSON RESPONSE TO ANDROGEN UP
| 86 | 61 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC DN
| 63 | 40 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION DN
| 87 | 57 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP
| 225 | 139 | All SZGR 2.0 genes in this pathway |
| LIAN LIPA TARGETS 6M
| 74 | 47 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART UP
| 103 | 69 | All SZGR 2.0 genes in this pathway |
| LIAN LIPA TARGETS 3M
| 59 | 36 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 2
| 72 | 52 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR UP
| 101 | 69 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2
| 163 | 113 | All SZGR 2.0 genes in this pathway |
| LI CYTIDINE ANALOGS CYCTOTOXICITY
| 15 | 8 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1
| 528 | 324 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4
| 307 | 185 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS
| 212 | 121 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN
| 409 | 268 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP
| 282 | 183 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN
| 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN
| 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN
| 591 | 366 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN
| 258 | 160 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA
| 51 | 35 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR UP
| 174 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP
| 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP
| 863 | 514 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN
| 940 | 425 | All SZGR 2.0 genes in this pathway |
| PIONTEK PKD1 TARGETS UP
| 38 | 24 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE D7 UP
| 107 | 67 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION A
| 67 | 52 | All SZGR 2.0 genes in this pathway |
| PARK APL PATHOGENESIS DN
| 50 | 35 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE
| 200 | 115 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN
| 277 | 166 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL DN
| 76 | 51 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 8
| 49 | 36 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP
| 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP
| 90 | 58 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR UP
| 55 | 41 | All SZGR 2.0 genes in this pathway |
| NOUSHMEHR GBM SILENCED BY METHYLATION
| 50 | 32 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP
| 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP
| 324 | 193 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN
| 918 | 550 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN
| 882 | 538 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN
| 321 | 200 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP
| 199 | 143 | All SZGR 2.0 genes in this pathway |
| KRIEG KDM3A TARGETS NOT HYPOXIA
| 208 | 107 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF
| 222 | 159 | All SZGR 2.0 genes in this pathway |
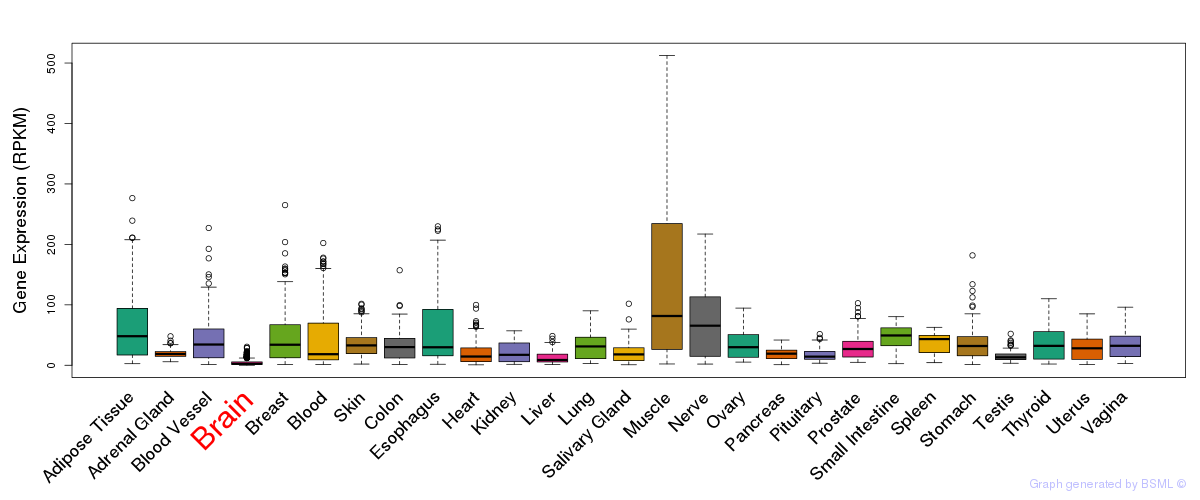
 eQTL annotation
eQTL annotation