Gene Page: SHANK2
Summary ?
| GeneID | 22941 |
| Symbol | SHANK2 |
| Synonyms | AUTS17|CORTBP1|CTTNBP1|ProSAP1|SHANK|SPANK-3 |
| Description | SH3 and multiple ankyrin repeat domains 2 |
| Reference | MIM:603290|HGNC:HGNC:14295|Ensembl:ENSG00000162105|HPRD:04479|Vega:OTTHUMG00000154615 |
| Gene type | protein-coding |
| Map location | 11q13.2 |
| Pascal p-value | 0.028 |
| Sherlock p-value | 0.191 |
| Fetal beta | 0.605 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | PROTEIN CLUSTERING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanNRC CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.061 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26025487 | 11 | 70674245 | SHANK2 | 1.175E-4 | 0.375 | 0.029 | DMG:Wockner_2014 |
| cg16456980 | 11 | 70714131 | SHANK2 | 2.874E-4 | -0.49 | 0.039 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16894557 | chr6 | 28999825 | SHANK2 | 22941 | 0.02 | trans |
Section II. Transcriptome annotation
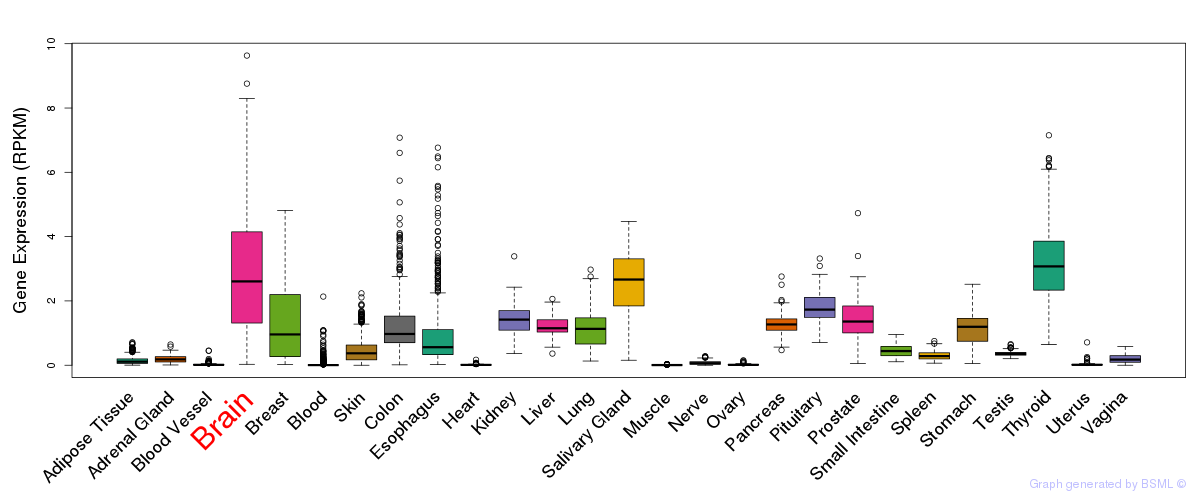
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ADNP | 0.95 | 0.96 |
| CYTSA | 0.95 | 0.96 |
| UBR5 | 0.94 | 0.95 |
| LATS1 | 0.94 | 0.96 |
| MGA | 0.94 | 0.94 |
| ZNF518B | 0.94 | 0.97 |
| NSD1 | 0.94 | 0.96 |
| MED1 | 0.94 | 0.96 |
| ARID2 | 0.94 | 0.95 |
| HMGXB4 | 0.94 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.70 | -0.88 |
| FXYD1 | -0.70 | -0.88 |
| HLA-F | -0.69 | -0.76 |
| AIFM3 | -0.69 | -0.76 |
| MT-CO2 | -0.69 | -0.88 |
| TSC22D4 | -0.68 | -0.79 |
| S100B | -0.68 | -0.82 |
| AF347015.27 | -0.68 | -0.84 |
| C5orf53 | -0.68 | -0.74 |
| IFI27 | -0.67 | -0.85 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0030160 | GKAP/Homer scaffold activity | NAS | Synap (GO term level: 6) | 10506216 |
| GO:0005515 | protein binding | IPI | 11583995 | |
| GO:0005515 | protein binding | NAS | - | |
| GO:0017124 | SH3 domain binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007242 | intracellular signaling cascade | NAS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARHGEF7 | BETA-PIX | COOL1 | DKFZp686C12170 | DKFZp761K1021 | KIAA0142 | KIAA0412 | Nbla10314 | P50 | P50BP | P85 | P85COOL1 | P85SPR | PAK3 | PIXB | Rho guanine nucleotide exchange factor (GEF) 7 | - | HPRD,BioGRID | 12626503 |
| BAI2 | - | brain-specific angiogenesis inhibitor 2 | - | HPRD,BioGRID | 10964907 |
| BAIAP2 | BAP2 | IRSP53 | BAI1-associated protein 2 | - | HPRD | 12504591 |
| CTTN | EMS1 | FLJ34459 | cortactin | - | HPRD,BioGRID | 9742101 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD,BioGRID | 10527873 |
| DLGAP1 | DAP-1 | DAP-1-ALPHA | DAP-1-BETA | GKAP | MGC88156 | SAPAP1 | hGKAP | discs, large (Drosophila) homolog-associated protein 1 | - | HPRD,BioGRID | 10527873 |
| DLGAP2 | DAP2 | SAPAP2 | discs, large (Drosophila) homolog-associated protein 2 | Two-hybrid | BioGRID | 10527873 |
| DLGAP3 | DAP3 | SAPAP3 | discs, large (Drosophila) homolog-associated protein 3 | Two-hybrid | BioGRID | 10527873 |
| DLGAP4 | DAP4 | KIAA0964 | MGC131862 | RP5-977B1.6 | SAPAP4 | discs, large (Drosophila) homolog-associated protein 4 | Two-hybrid | BioGRID | 10527873 |
| DNM2 | CMTDI1 | CMTDIB | DI-CMTB | DYN2 | DYNII | dynamin 2 | - | HPRD,BioGRID | 11583995 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | Affinity Capture-Western | BioGRID | 10844022 |
| DYNLL2 | DNCL1B | Dlc2 | MGC17810 | dynein, light chain, LC8-type 2 | Affinity Capture-Western | BioGRID | 10844022 |
| LPHN1 | CIRL1 | CL1 | LEC2 | latrophilin 1 | - | HPRD,BioGRID | 10964907 |
| LPHN2 | CIRL2 | CL2 | LEC1 | LPHH1 | latrophilin 2 | - | HPRD,BioGRID | 10964907 |
| MYO5A | GS1 | MYH12 | MYO5 | MYR12 | myosin VA (heavy chain 12, myoxin) | Affinity Capture-Western | BioGRID | 10844022 |
| SSTR2 | - | somatostatin receptor 2 | - | HPRD | 10373412 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SCHUETZ BREAST CANCER DUCTAL INVASIVE DN | 84 | 53 | All SZGR 2.0 genes in this pathway |
| FRASOR TAMOXIFEN RESPONSE UP | 51 | 36 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA DN | 77 | 52 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS UP | 82 | 58 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 11Q12 Q14 AMPLICON | 158 | 93 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD DN | 150 | 93 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY DN | 145 | 88 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| MEISSNER ES ICP WITH H3K4ME3 | 34 | 17 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC ICP WITH H3 UNMETHYLATED | 24 | 15 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN ICP WITH H3K4ME3 | 32 | 17 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 ICP WITH H3K27ME3 | 74 | 46 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA1 DN | 74 | 47 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS DN | 116 | 74 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 2883 | 2889 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| hsa-miR-101 | UACAGUACUGUGAUAACUGAAG | ||||
| miR-124/506 | 4847 | 4853 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA | ||||
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-130/301 | 3358 | 3364 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-137 | 2716 | 2723 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-142-5p | 3326 | 3333 | 1A,m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-144 | 3404 | 3410 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG | ||||
| miR-186 | 1673 | 1679 | 1A | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-19 | 3357 | 3363 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-218 | 1713 | 1720 | 1A,m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU | ||||
| miR-221/222 | 4824 | 4830 | m8 | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-23 | 3045 | 3051 | m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-323 | 3045 | 3051 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-324-3p | 648 | 654 | m8 | hsa-miR-324-3p | CCACUGCCCCAGGUGCUGCUGG |
| miR-33 | 4879 | 4885 | m8 | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-374 | 1574 | 1580 | 1A | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-381 | 3385 | 3391 | m8 | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-495 | 2874 | 2880 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-7 | 2293 | 2300 | 1A,m8 | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
| miR-9 | 127 | 133 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.