| KEGG MAPK SIGNALING PATHWAY
| 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION
| 201 | 138 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES
| 66 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING
| 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM
| 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM
| 270 | 204 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA DN
| 116 | 79 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN UP
| 184 | 125 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP
| 290 | 177 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN
| 158 | 102 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D UP
| 194 | 122 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D UP
| 175 | 108 | All SZGR 2.0 genes in this pathway |
| LOPEZ MESOTELIOMA SURVIVAL TIME UP
| 14 | 8 | All SZGR 2.0 genes in this pathway |
| BILBAN B CLL LPL UP
| 63 | 39 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP
| 204 | 140 | All SZGR 2.0 genes in this pathway |
| LOPEZ MESOTHELIOMA SURVIVAL WORST VS BEST UP
| 14 | 10 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN
| 329 | 219 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP
| 171 | 112 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP
| 265 | 158 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN
| 805 | 505 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA FOREVER DN
| 31 | 23 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 UP
| 150 | 93 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP
| 536 | 340 | All SZGR 2.0 genes in this pathway |
| BEGUM TARGETS OF PAX3 FOXO1 FUSION DN
| 45 | 34 | All SZGR 2.0 genes in this pathway |
| DAUER STAT3 TARGETS UP
| 49 | 35 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN
| 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN
| 483 | 336 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP
| 195 | 138 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES UP
| 27 | 19 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN
| 464 | 276 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP
| 398 | 262 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS
| 882 | 572 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA LB UP
| 48 | 28 | All SZGR 2.0 genes in this pathway |
| CROMER METASTASIS DN
| 81 | 58 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN
| 246 | 180 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C8
| 72 | 56 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS UP
| 56 | 35 | All SZGR 2.0 genes in this pathway |
| KANG FLUOROURACIL RESISTANCE DN
| 16 | 10 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 8
| 86 | 57 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS UP
| 85 | 50 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR
| 681 | 420 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE
| 261 | 166 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1
| 528 | 324 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4
| 307 | 185 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN
| 409 | 268 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP
| 461 | 298 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN
| 258 | 160 | All SZGR 2.0 genes in this pathway |
| MASSARWEH RESPONSE TO ESTRADIOL
| 61 | 47 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS DN
| 145 | 93 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 DN
| 78 | 49 | All SZGR 2.0 genes in this pathway |
| PARK TRETINOIN RESPONSE AND RARA PLZF FUSION
| 22 | 14 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN
| 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN
| 668 | 419 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 10
| 33 | 22 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN
| 134 | 83 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION
| 130 | 87 | All SZGR 2.0 genes in this pathway |
| LOPEZ MESOTHELIOMA SURVIVAL OVERALL DN
| 16 | 8 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2
| 277 | 172 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S2
| 115 | 74 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE
| 721 | 492 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS UP
| 127 | 78 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION
| 456 | 287 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP
| 229 | 149 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP
| 211 | 131 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP
| 281 | 183 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP
| 430 | 288 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN
| 1080 | 713 | All SZGR 2.0 genes in this pathway |
| GUO TARGETS OF IRS1 AND IRS2
| 98 | 67 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP
| 199 | 143 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN
| 784 | 464 | All SZGR 2.0 genes in this pathway |
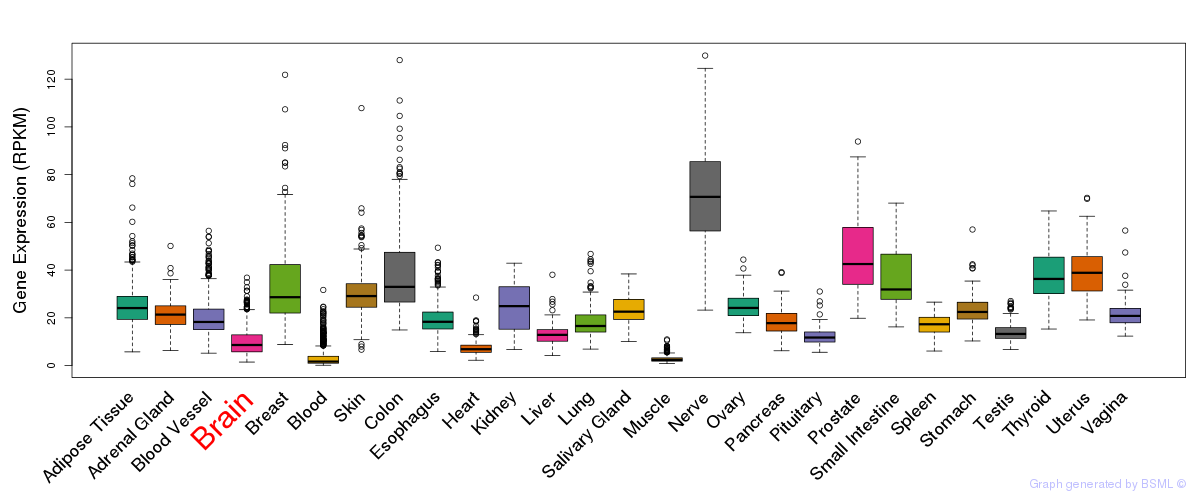
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation