Gene Page: SF3B1
Summary ?
| GeneID | 23451 |
| Symbol | SF3B1 |
| Synonyms | Hsh155|MDS|PRP10|PRPF10|SAP155|SF3b155 |
| Description | splicing factor 3b subunit 1 |
| Reference | MIM:605590|HGNC:HGNC:10768|Ensembl:ENSG00000115524|HPRD:09281|Vega:OTTHUMG00000154447 |
| Gene type | protein-coding |
| Map location | 2q33.1 |
| Pascal p-value | 1.471E-10 |
| Sherlock p-value | 0.355 |
| Fetal beta | 0.888 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:PGC128 | Genome-wide Association Study | A multi-stage schizophrenia GWAS of up to 36,989 cases and 113,075 controls. Reported by the Schizophrenia Working Group of PGC. 128 independent associations spanning 108 loci | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 1.907 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 CV:PGC128
CV:PGC128
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs6434928 | chr2 | 198304577 | AG | 1.478E-11 | intergenic | SF3B1,COQ10B | dist=4760;dist=13654 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06750227 | 2 | 198298917 | SF3B1 | 2.37E-8 | -0.017 | 7.79E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
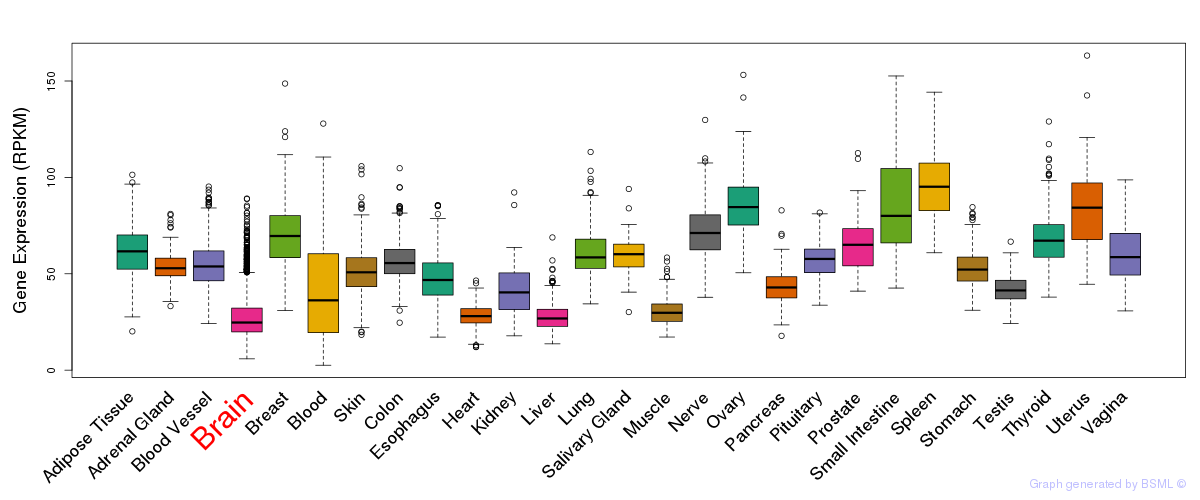
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003682 | chromatin binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 17353931 | |
| GO:0031202 | RNA splicing factor activity, transesterification mechanism | NAS | 9585501 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000398 | nuclear mRNA splicing, via spliceosome | EXP | 12226669 | |
| GO:0000398 | nuclear mRNA splicing, via spliceosome | NAS | 9585501 | |
| GO:0008380 | RNA splicing | IEA | - | |
| GO:0009952 | anterior/posterior pattern formation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005681 | spliceosome | NAS | 9585501 | |
| GO:0016607 | nuclear speck | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATG12 | APG12 | APG12L | FBR93 | HAPG12 | ATG12 autophagy related 12 homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| BRF2 | BRFU | FLJ11052 | TFIIIB50 | BRF2, subunit of RNA polymerase III transcription initiation factor, BRF1-like | Affinity Capture-MS | BioGRID | 17353931 |
| CCNE1 | CCNE | cyclin E1 | Affinity Capture-Western | BioGRID | 9891079 |
| DDX42 | FLJ43179 | RHELP | RNAHP | SF3b125 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 42 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 12234937 |
| GSK3B | - | glycogen synthase kinase 3 beta | Affinity Capture-MS | BioGRID | 17353931 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | Affinity Capture-MS | BioGRID | 17353931 |
| PHF5A | INI | MGC1346 | SF3b14b | bK223H9.2 | PHD finger protein 5A | - | HPRD,BioGRID | 12234937 |
| PPP1R8 | ARD-1 | ARD1 | NIPP-1 | NIPP1 | PRO2047 | protein phosphatase 1, regulatory (inhibitor) subunit 8 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12105215 |
| PUF60 | FIR | FLJ31379 | RoBPI | SIAHBP1 | poly-U binding splicing factor 60KDa | Affinity Capture-MS | BioGRID | 17353931 |
| RNPS1 | E5.1 | MGC117332 | RNA binding protein S1, serine-rich domain | Affinity Capture-MS | BioGRID | 17353931 |
| RNU11 | U11 | RNA, U11 small nuclear | Affinity Capture-MS | BioGRID | 15146077 |
| RNU12 | U12 | RNA, U12 small nuclear | Affinity Capture-MS | BioGRID | 15146077 |
| SF3A2 | PRP11 | PRPF11 | SAP62 | SF3a66 | splicing factor 3a, subunit 2, 66kDa | Affinity Capture-MS | BioGRID | 12234937 |
| SF3B14 | CGI-110 | HSPC175 | Ht006 | P14 | SAP14 | SF3B14a | splicing factor 3B, 14 kDa subunit | - | HPRD,BioGRID | 11500380 |
| SF3B14 | CGI-110 | HSPC175 | Ht006 | P14 | SAP14 | SF3B14a | splicing factor 3B, 14 kDa subunit | p14 interacts with SF3b155. | BIND | 11500380 |
| SF3B2 | SAP145 | SF3B145 | SF3b1 | SF3b150 | splicing factor 3b, subunit 2, 145kDa | - | HPRD,BioGRID | 10490618 |
| SF3B3 | KIAA0017 | RSE1 | SAP130 | SF3b130 | STAF130 | splicing factor 3b, subunit 3, 130kDa | - | HPRD | 10490618 |11406595 |12234937 |
| SF3B3 | KIAA0017 | RSE1 | SAP130 | SF3b130 | STAF130 | splicing factor 3b, subunit 3, 130kDa | Affinity Capture-MS Affinity Capture-Western | BioGRID | 10490618 |12234937 |
| SF3B4 | MGC10828 | SAP49 | SF3b49 | splicing factor 3b, subunit 4, 49kDa | Affinity Capture-MS | BioGRID | 12234937 |
| SF3B5 | MGC3133 | SF3b10 | splicing factor 3b, subunit 5, 10kDa | Affinity Capture-MS | BioGRID | 12234937 |
| SMNDC1 | SMNR | SPF30 | survival motor neuron domain containing 1 | Affinity Capture-MS | BioGRID | 17353931 |
| SNRNP40 | 40K | FLJ41108 | HPRP8BP | MGC1910 | PRP8BP | PRPF8BP | RP11-490K7.3 | SPF38 | WDR57 | small nuclear ribonucleoprotein 40kDa (U5) | Affinity Capture-MS | BioGRID | 17353931 |
| TCERG1 | CA150 | MGC133200 | TAF2S | transcription elongation regulator 1 | - | HPRD,BioGRID | 15456888 |
| U2AF1 | DKFZp313J1712 | FP793 | RN | RNU2AF1 | U2AF35 | U2AFBP | U2 small nuclear RNA auxiliary factor 1 | SAP155 interacts with U2AF35. | BIND | 9671485 |
| U2AF2 | U2AF65 | U2 small nuclear RNA auxiliary factor 2 | SAP155 interacts with U2AF65. | BIND | 9671485 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING | 111 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING MINOR PATHWAY | 45 | 19 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED UP | 78 | 48 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| BERENJENO ROCK SIGNALING NOT VIA RHOA DN | 48 | 34 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| WOOD EBV EBNA1 TARGETS UP | 110 | 71 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BYSTROEM CORRELATED WITH IL5 DN | 64 | 47 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR1 TARGETS UP | 53 | 39 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-204/211 | 282 | 289 | 1A,m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-493-5p | 315 | 321 | 1A | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-543 | 287 | 293 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.