Gene Page: SNAPIN
Summary ?
| GeneID | 23557 |
| Symbol | SNAPIN |
| Synonyms | BLOC1S7|BLOS7|BORCS3|SNAPAP |
| Description | SNAP associated protein |
| Reference | MIM:607007|HGNC:HGNC:17145|Ensembl:ENSG00000143553|HPRD:06111|Vega:OTTHUMG00000037086 |
| Gene type | protein-coding |
| Map location | 1q21.3 |
| Sherlock p-value | 0.836 |
| Fetal beta | 0.245 |
| Support | CANABINOID DOPAMINE EXOCYTOSIS SEROTONIN |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 9.7664 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
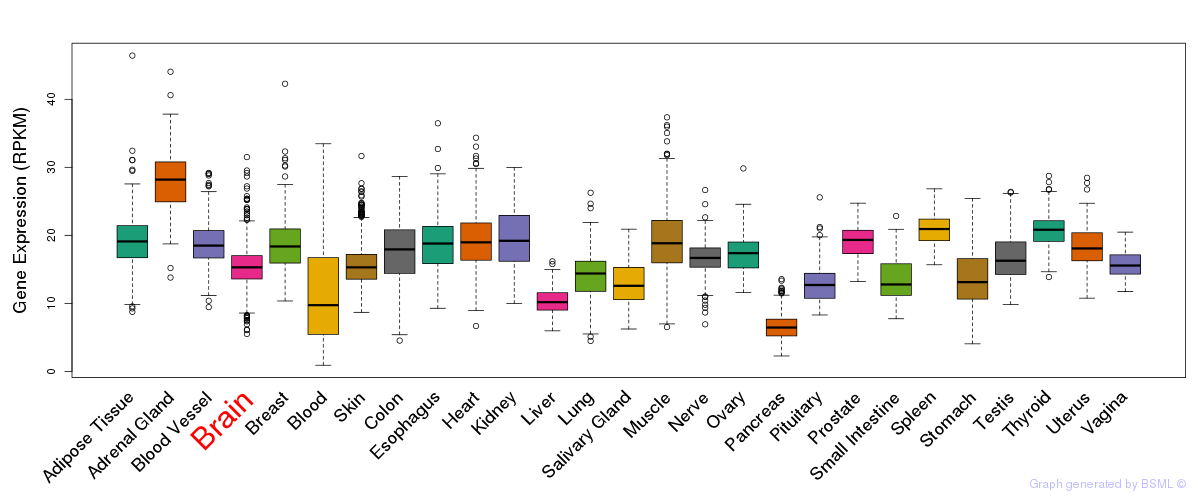
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000149 | SNARE binding | TAS | 10195194 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007269 | neurotransmitter secretion | TAS | neuron, axon, Synap, serotonin, Neurotransmitter, dopamine (GO term level: 8) | 10195194 |
| GO:0006886 | intracellular protein transport | TAS | 10195194 | |
| GO:0006887 | exocytosis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008021 | synaptic vesicle | TAS | Synap, Neurotransmitter (GO term level: 12) | 10195194 |
| GO:0019717 | synaptosome | IEA | Synap, Brain (GO term level: 7) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - | |
| GO:0031410 | cytoplasmic vesicle | IEA | - | |
| GO:0030659 | cytoplasmic vesicle membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BLOC1S1 | BLOS1 | FLJ39337 | FLJ97089 | GCN5L1 | MGC87455 | MICoA | RT14 | biogenesis of lysosomal organelles complex-1, subunit 1 | Snapin interacts with BLOS1. | BIND | 15102850 |
| BLOC1S1 | BLOS1 | FLJ39337 | FLJ97089 | GCN5L1 | MGC87455 | MICoA | RT14 | biogenesis of lysosomal organelles complex-1, subunit 1 | Two-hybrid | BioGRID | 15102850 |
| BLOC1S2 | BLOS2 | FLJ30135 | MGC10120 | RP11-316M21.4 | biogenesis of lysosomal organelles complex-1, subunit 2 | - | HPRD,BioGRID | 15102850 |
| BLOC1S2 | BLOS2 | FLJ30135 | MGC10120 | RP11-316M21.4 | biogenesis of lysosomal organelles complex-1, subunit 2 | Snapin interacts with BLOS2. | BIND | 15102850 |
| BLOC1S3 | BLOS3 | FLJ26641 | FLJ26676 | HPS8 | RP | biogenesis of lysosomal organelles complex-1, subunit 3 | - | HPRD,BioGRID | 15102850 |
| CNO | BCAS4L | FLJ11230 | cappuccino homolog (mouse) | Snapin interacts with Cappuccino. | BIND | 15102850 |
| CNO | BCAS4L | FLJ11230 | cappuccino homolog (mouse) | - | HPRD,BioGRID | 15102850 |
| DDR1 | CAK | CD167 | DDR | EDDR1 | MCK10 | NEP | NTRK4 | PTK3 | PTK3A | RTK6 | TRKE | discoidin domain receptor tyrosine kinase 1 | Two-hybrid | BioGRID | 16169070 |
| DTNBP1 | DBND | DKFZp564K192 | FLJ30031 | HPS7 | MGC20210 | My031 | SDY | dystrobrevin binding protein 1 | Affinity Capture-Western Two-hybrid | BioGRID | 15102850 |
| DTNBP1 | DBND | DKFZp564K192 | FLJ30031 | HPS7 | MGC20210 | My031 | SDY | dystrobrevin binding protein 1 | Snapin interacts with Dysbindin. | BIND | 15102850 |
| EBAG9 | EB9 | PDAF | RCAS1 | estrogen receptor binding site associated, antigen, 9 | EBAG9 interacts with Snapin. | BIND | 15635093 |
| EEF1G | EF1G | GIG35 | eukaryotic translation elongation factor 1 gamma | Two-hybrid | BioGRID | 16169070 |
| GPRASP1 | GASP | GASP1 | KIAA0443 | G protein-coupled receptor associated sorting protein 1 | Two-hybrid | BioGRID | 16169070 |
| HAP1 | HAP2 | HIP5 | HLP | hHLP1 | huntingtin-associated protein 1 | Two-hybrid | BioGRID | 16169070 |
| IKBKAP | DKFZp781H1425 | DYS | ELP1 | FD | FLJ12497 | IKAP | IKI3 | TOT1 | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein | Two-hybrid | BioGRID | 16169070 |
| IMMT | DKFZp779P1653 | HMP | MGC111146 | P87 | P87/89 | P89 | PIG4 | PIG52 | inner membrane protein, mitochondrial (mitofilin) | Two-hybrid | BioGRID | 16169070 |
| KAT5 | ESA1 | HTATIP | HTATIP1 | PLIP | TIP | TIP60 | cPLA2 | K(lysine) acetyltransferase 5 | Two-hybrid | BioGRID | 16169070 |
| KIF5C | FLJ44735 | KIAA0531 | KINN | MGC111478 | NKHC | NKHC-2 | NKHC2 | kinesin family member 5C | Two-hybrid | BioGRID | 16169070 |
| LAMC1 | LAMB2 | MGC87297 | laminin, gamma 1 (formerly LAMB2) | Two-hybrid | BioGRID | 16169070 |
| LRP12 | DKFZp781F1053 | FLJ12929 | ST7 | low density lipoprotein-related protein 12 | - | HPRD | 12809483 |
| MYST2 | HBO1 | HBOA | KAT7 | MYST histone acetyltransferase 2 | Two-hybrid | BioGRID | 16169070 |
| PLDN | PA | PALLID | pallidin homolog (mouse) | - | HPRD,BioGRID | 15102850 |
| RGS7 | - | regulator of G-protein signaling 7 | - | HPRD,BioGRID | 12659861 |
| SNAP23 | HsT17016 | SNAP23A | SNAP23B | synaptosomal-associated protein, 23kDa | SNAP23 interacts with Snapin. | BIND | 15635093 |
| SNAP23 | HsT17016 | SNAP23A | SNAP23B | synaptosomal-associated protein, 23kDa | - | HPRD,BioGRID | 12877659 |
| SNAP25 | FLJ23079 | RIC-4 | RIC4 | SEC9 | SNAP | SNAP-25 | bA416N4.2 | dJ1068F16.2 | synaptosomal-associated protein, 25kDa | - | HPRD,BioGRID | 10195194 |
| SNAP25 | FLJ23079 | RIC-4 | RIC4 | SEC9 | SNAP | SNAP-25 | bA416N4.2 | dJ1068F16.2 | synaptosomal-associated protein, 25kDa | SNAP25 interacts with Snapin. | BIND | 15635093 |
| SPAG5 | DEEPEST | MAP126 | hMAP126 | sperm associated antigen 5 | Two-hybrid | BioGRID | 16169070 |
| ST7 | DKFZp762O2113 | ETS7q | FAM4A | FAM4A1 | HELG | RAY1 | SEN4 | TSG7 | suppression of tumorigenicity 7 | Two-hybrid | BioGRID | 12809483 |
| TRPV1 | DKFZp434K0220 | VR1 | transient receptor potential cation channel, subfamily V, member 1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 15066994 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME MEMBRANE TRAFFICKING | 129 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | 60 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | 53 | 27 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| MORI LARGE PRE BII LYMPHOCYTE DN | 58 | 39 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 11 | 57 | 40 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |