Gene Page: POLA2
Summary ?
| GeneID | 23649 |
| Symbol | POLA2 |
| Synonyms | - |
| Description | polymerase (DNA) alpha 2, accessory subunit |
| Reference | HGNC:HGNC:30073|Ensembl:ENSG00000014138|HPRD:10160|Vega:OTTHUMG00000165951 |
| Gene type | protein-coding |
| Map location | 11q13.1 |
| Pascal p-value | 0.157 |
| Sherlock p-value | 0.412 |
| Fetal beta | 0.34 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cortex Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21257117 | 11 | 65029643 | POLA2 | 2.45E-8 | -0.011 | 7.95E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
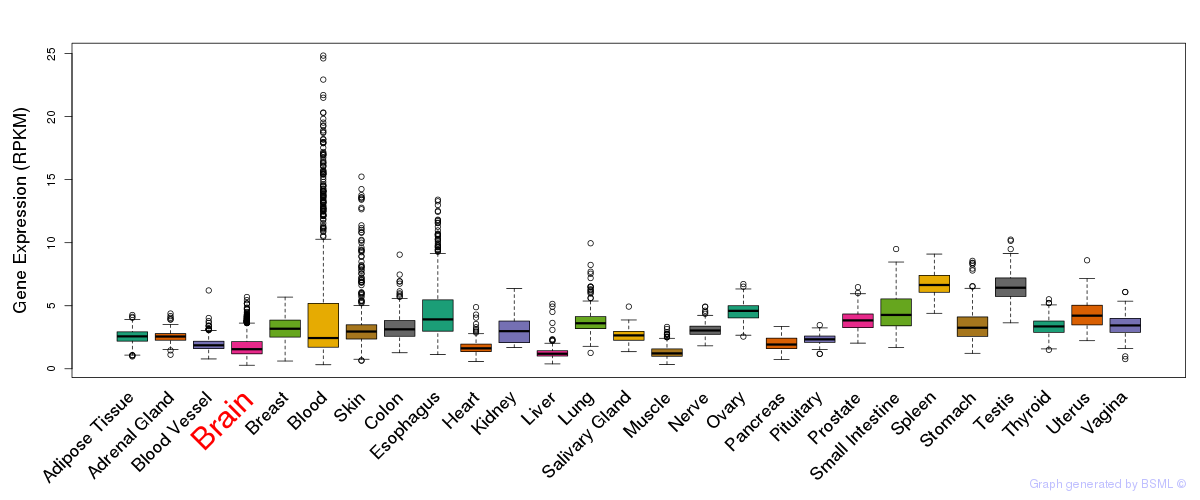
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKTIP | FT1 | FTS | AKT interacting protein | Two-hybrid | BioGRID | 16169070 |
| ASCC2 | ASC1p100 | activating signal cointegrator 1 complex subunit 2 | Two-hybrid | BioGRID | 16169070 |
| ATP5C1 | ATP5C | ATP5CL1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | Two-hybrid | BioGRID | 16169070 |
| C8orf30A | FLJ40907 | chromosome 8 open reading frame 30A | Two-hybrid | BioGRID | 16169070 |
| C9orf25 | FLJ39031 | bA573M23.5 | chromosome 9 open reading frame 25 | Two-hybrid | BioGRID | 16169070 |
| CPE | - | carboxypeptidase E | Two-hybrid | BioGRID | 16169070 |
| CRELD1 | AVSD2 | CIRRIN | DKFZp566D213 | cysteine-rich with EGF-like domains 1 | Two-hybrid | BioGRID | 16169070 |
| DDX24 | - | DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 | Two-hybrid | BioGRID | 16169070 |
| DHX34 | DDX34 | HRH1 | KIAA0134 | DEAH (Asp-Glu-Ala-His) box polypeptide 34 | Two-hybrid | BioGRID | 16169070 |
| EIF6 | 2 | CAB | EIF3A | ITGB4BP | b | b(2)gcn | gcn | p27BBP | eukaryotic translation initiation factor 6 | Two-hybrid | BioGRID | 16169070 |
| EZH2 | ENX-1 | EZH1 | KMT6 | MGC9169 | enhancer of zeste homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| FGFR3 | ACH | CD333 | CEK2 | HSFGFR3EX | JTK4 | fibroblast growth factor receptor 3 | Two-hybrid | BioGRID | 16169070 |
| HELB | - | helicase (DNA) B | - | HPRD,BioGRID | 12181327 |
| IGFBP3 | BP-53 | IBP3 | insulin-like growth factor binding protein 3 | Two-hybrid | BioGRID | 16169070 |
| KLF6 | BCD1 | COPEB | CPBP | DKFZp686N0199 | GBF | PAC1 | ST12 | ZF9 | Kruppel-like factor 6 | Two-hybrid | BioGRID | 16169070 |
| LRPAP1 | A2MRAP | A2RAP | HBP44 | MGC138272 | MRAP | RAP | low density lipoprotein receptor-related protein associated protein 1 | Two-hybrid | BioGRID | 16169070 |
| NHP2L1 | 15.5K | FA-1 | FA1 | NHPX | OTK27 | SNU13 | SPAG12 | SSFA1 | NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| PARP1 | ADPRT | ADPRT1 | PARP | PARP-1 | PPOL | pADPRT-1 | poly (ADP-ribose) polymerase 1 | - | HPRD,BioGRID | 9518481 |
| POLA1 | DKFZp686K1672 | POLA | p180 | polymerase (DNA directed), alpha 1, catalytic subunit | - | HPRD,BioGRID | 8223465 |12546699 |
| PRIM1 | MGC12308 | p49 | primase, DNA, polypeptide 1 (49kDa) | - | HPRD | 9705292 |
| PRIM2 | MGC75142 | PRIM2A | p58 | primase, DNA, polypeptide 2 (58kDa) | - | HPRD | 9705292 |
| PRPF38A | FLJ14936 | MGC3320 | RP5-965L7.1 | PRP38 pre-mRNA processing factor 38 (yeast) domain containing A | Two-hybrid | BioGRID | 16169070 |
| RBM23 | CAPERbeta | FLJ10482 | MGC4458 | PP239 | RNPC4 | RNA binding motif protein 23 | Two-hybrid | BioGRID | 16169070 |
| SETDB1 | ESET | KG1T | KIAA0067 | KMT1E | SET domain, bifurcated 1 | Two-hybrid | BioGRID | 16169070 |
| STK40 | MGC4796 | RP11-268J15.4 | SHIK | SgK495 | serine/threonine kinase 40 | Two-hybrid | BioGRID | 16169070 |
| ULK2 | KIAA0623 | Unc51.2 | unc-51-like kinase 2 (C. elegans) | Two-hybrid | BioGRID | 16169070 |
| XPNPEP1 | SAMP | XPNPEP | XPNPEPL | XPNPEPL1 | X-prolyl aminopeptidase (aminopeptidase P) 1, soluble | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PURINE METABOLISM | 159 | 96 | All SZGR 2.0 genes in this pathway |
| KEGG PYRIMIDINE METABOLISM | 98 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG DNA REPLICATION | 36 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | 31 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME M G1 TRANSITION | 81 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 S TRANSITION | 112 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME POL SWITCHING | 13 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF DNA | 92 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME CHROMOSOME MAINTENANCE | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME LAGGING STRAND SYNTHESIS | 19 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | 13 | 6 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME E2F MEDIATED REGULATION OF DNA REPLICATION | 35 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TELOMERE MAINTENANCE | 75 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME EXTENSION OF TELOMERES | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME S PHASE | 109 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA STRAND ELONGATION | 30 | 18 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| ROSTY CERVICAL CANCER PROLIFERATION CLUSTER | 140 | 73 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC 8HR 3 DN | 10 | 7 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| REN BOUND BY E2F | 61 | 40 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION DN | 87 | 57 | All SZGR 2.0 genes in this pathway |
| KANG DOXORUBICIN RESISTANCE UP | 54 | 33 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| LEE AGING MUSCLE DN | 46 | 29 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| KALMA E2F1 TARGETS | 11 | 7 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP | 202 | 115 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 16HR | 40 | 24 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO SERM OR FULVESTRANT DN | 50 | 29 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO METHOTREXATE | 18 | 9 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |