Gene Page: ALK
Summary ?
| GeneID | 238 |
| Symbol | ALK |
| Synonyms | CD246|NBLST3 |
| Description | anaplastic lymphoma receptor tyrosine kinase |
| Reference | MIM:105590|HGNC:HGNC:427|Ensembl:ENSG00000171094|HPRD:00104|Vega:OTTHUMG00000152034 |
| Gene type | protein-coding |
| Map location | 2p23 |
| Pascal p-value | 0.015 |
| Sherlock p-value | 0.825 |
| Fetal beta | -0.495 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
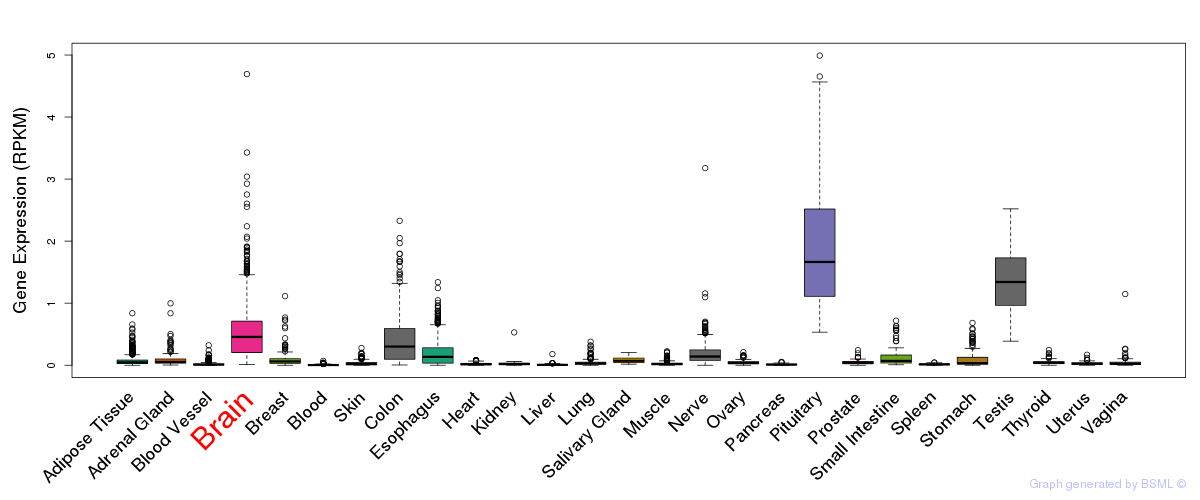
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TBXAS1 | 0.87 | 0.83 |
| C3AR1 | 0.86 | 0.80 |
| P2RY12 | 0.81 | 0.79 |
| C1QC | 0.81 | 0.83 |
| LAPTM5 | 0.80 | 0.84 |
| ADORA3 | 0.80 | 0.82 |
| RNASE6 | 0.78 | 0.82 |
| CD53 | 0.77 | 0.80 |
| C3 | 0.77 | 0.82 |
| GPR34 | 0.77 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SH3BP2 | -0.34 | -0.42 |
| ZNF814 | -0.34 | -0.40 |
| ZNF418 | -0.34 | -0.31 |
| KIAA1949 | -0.33 | -0.30 |
| VAV2 | -0.33 | -0.27 |
| GJC1 | -0.33 | -0.32 |
| AC004017.1 | -0.33 | -0.25 |
| AL033532.1 | -0.33 | -0.26 |
| AC010522.1 | -0.33 | -0.39 |
| UPF3A | -0.33 | -0.39 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004714 | transmembrane receptor protein tyrosine kinase activity | IDA | neurite (GO term level: 8) | 9174053 |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004716 | receptor signaling protein tyrosine kinase activity | TAS | 9053841 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | TAS | Brain (GO term level: 7) | 9053841 |
| GO:0007399 | nervous system development | IMP | neurite (GO term level: 5) | 11121404 |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0006487 | protein amino acid N-linked glycosylation | IMP | 9174053 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 9053841 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DAVICIONI PAX FOXO1 SIGNATURE IN ARMS UP | 59 | 38 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| DAVICIONI RHABDOMYOSARCOMA PAX FOXO1 FUSION UP | 64 | 37 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS DN | 123 | 78 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D DN | 143 | 83 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 UP | 276 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 2 UP | 139 | 83 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA COAMPLIFIED WITH MYCN | 43 | 29 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| CHEN NEUROBLASTOMA COPY NUMBER GAINS | 50 | 35 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| TAVAZOIE METASTASIS | 108 | 68 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-96 | 107 | 113 | m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.