Gene Page: PTF1A
Summary ?
| GeneID | 256297 |
| Symbol | PTF1A |
| Synonyms | PACA|PAGEN2|PTF1-p48|bHLHa29 |
| Description | pancreas specific transcription factor, 1a |
| Reference | MIM:607194|HGNC:HGNC:23734|Ensembl:ENSG00000168267|HPRD:09526|Vega:OTTHUMG00000017815 |
| Gene type | protein-coding |
| Map location | 10p12.2 |
| Pascal p-value | 0.196 |
| Fetal beta | -0.394 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18759960 | 10 | 23481776 | PTF1A | 2.72E-5 | -0.249 | 0.018 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
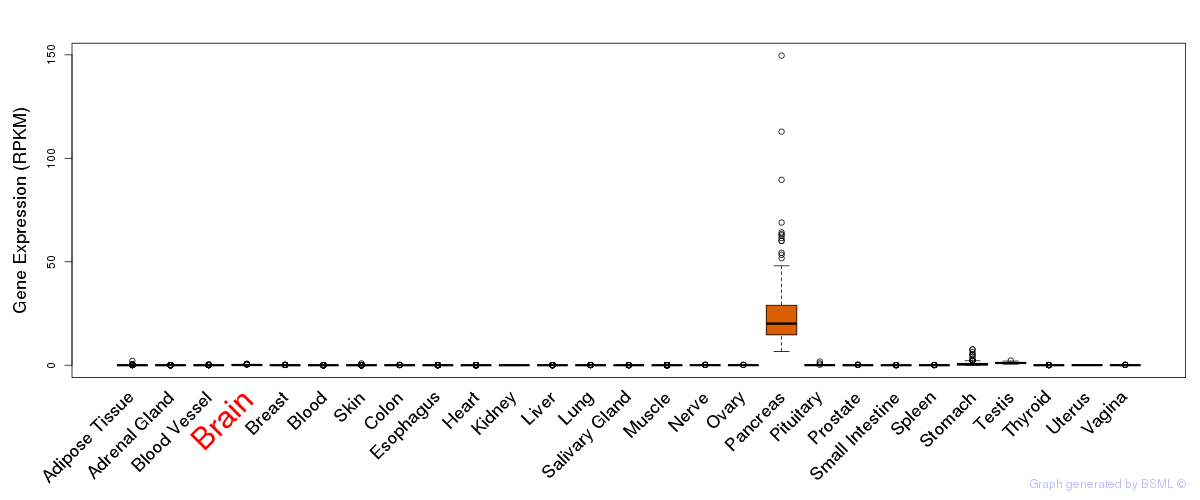
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | ISS | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0030528 | transcription regulator activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0048699 | generation of neurons | IEA | neuron, neurogenesis (GO term level: 7) | - |
| GO:0048663 | neuron fate commitment | IEA | neuron (GO term level: 9) | - |
| GO:0030902 | hindbrain development | IEA | Brain (GO term level: 8) | - |
| GO:0006355 | regulation of transcription, DNA-dependent | ISS | - | |
| GO:0009790 | embryonic development | IEA | - | |
| GO:0009888 | tissue development | IDA | 12185368 | |
| GO:0031017 | exocrine pancreas development | ISS | - | |
| GO:0048384 | retinoic acid receptor signaling pathway | IEA | - | |
| GO:0045941 | positive regulation of transcription | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | ISS | - | |
| GO:0005667 | transcription factor complex | ISS | - | |
| GO:0005737 | cytoplasm | ISS | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID HES HEY PATHWAY | 48 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF BETA CELL DEVELOPMENT | 30 | 23 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| ZHOU PANCREATIC EXOCRINE PROGENITOR | 11 | 10 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-410 | 182 | 188 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.