Gene Page: UPF2
Summary ?
| GeneID | 26019 |
| Symbol | UPF2 |
| Synonyms | HUPF2|RENT2|smg-3 |
| Description | UPF2 regulator of nonsense transcripts homolog (yeast) |
| Reference | MIM:605529|HGNC:HGNC:17854|Ensembl:ENSG00000151461|HPRD:10405|Vega:OTTHUMG00000017678 |
| Gene type | protein-coding |
| Map location | 10p14-p13 |
| Pascal p-value | 0.811 |
| Sherlock p-value | 0.722 |
| Fetal beta | -0.172 |
| DMG | 1 (# studies) |
| eGene | Meta |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| DNM:Gulsuner_2013 | Whole Exome Sequencing analysis | 155 DNMs identified by exome sequencing of quads or trios of schizophrenia individuals and their parents. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UPF2 | chr10 | 12041981 | C | A | NM_015542 NM_080599 | p.561G>V p.561G>V | missense missense | Schizophrenia | DNM:Gulsuner_2013 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25858261 | 10 | 12085058 | UPF2 | 3.16E-8 | -0.011 | 9.57E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
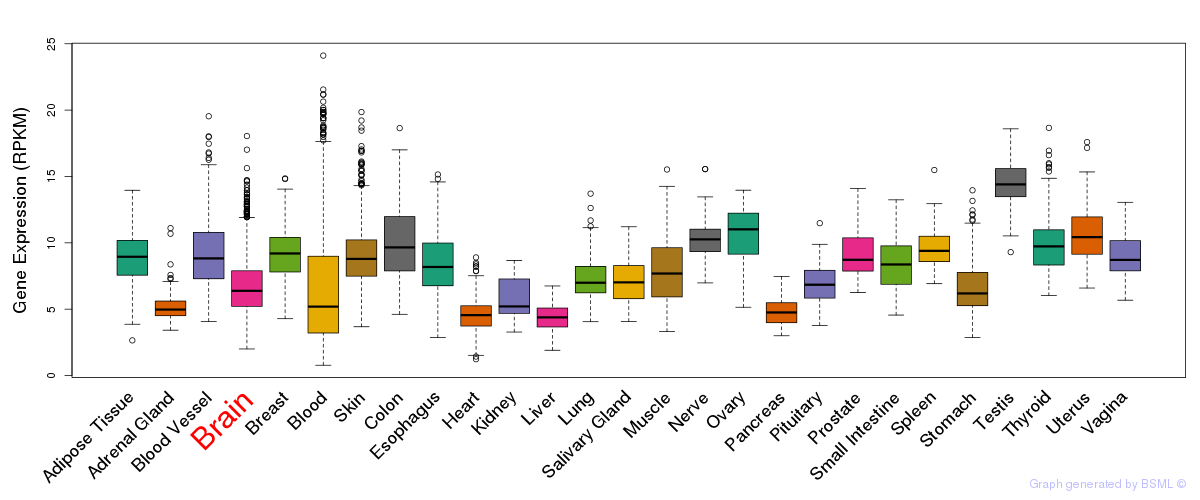
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MELK | 0.97 | 0.83 |
| TTK | 0.97 | 0.86 |
| PBK | 0.97 | 0.75 |
| BUB1 | 0.96 | 0.82 |
| DLGAP5 | 0.96 | 0.79 |
| NDC80 | 0.96 | 0.72 |
| CEP55 | 0.96 | 0.80 |
| KIF23 | 0.96 | 0.84 |
| SGOL1 | 0.96 | 0.73 |
| ESCO2 | 0.96 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.46 | -0.76 |
| AF347015.27 | -0.44 | -0.80 |
| C5orf53 | -0.44 | -0.66 |
| PTH1R | -0.44 | -0.69 |
| AF347015.31 | -0.44 | -0.79 |
| AF347015.33 | -0.44 | -0.81 |
| MT-CO2 | -0.44 | -0.82 |
| FBXO2 | -0.44 | -0.62 |
| AIFM3 | -0.44 | -0.71 |
| TINAGL1 | -0.43 | -0.72 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003723 | RNA binding | IEA | - | |
| GO:0042802 | identical protein binding | IPI | 15231747 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | TAS | 16488880 | |
| GO:0006406 | mRNA export from nucleus | TAS | 16488880 | |
| GO:0016070 | RNA metabolic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 14636577 | |
| GO:0005737 | cytoplasm | IDA | 14636577 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| DCP1A | FLJ21691 | HSA275986 | Nbla00360 | SMAD4IP1 | SMIF | DCP1 decapping enzyme homolog A (S. cerevisiae) | - | HPRD | 12417715 |
| DCP2 | FLJ33245 | NUDT20 | DCP2 decapping enzyme homolog (S. cerevisiae) | Affinity Capture-Western | BioGRID | 14527413 |
| EIF1 | A121 | EIF-1 | EIF1A | ISO1 | SUI1 | eukaryotic translation initiation factor 1 | - | HPRD | 11073994 |
| EIF4A1 | DDX2A | EIF-4A | EIF4A | eukaryotic translation initiation factor 4A, isoform 1 | - | HPRD,BioGRID | 11073994 |
| EXOSC10 | PM-Scl | PM/Scl-100 | PMSCL | PMSCL2 | RRP6 | Rrp6p | p2 | p3 | p4 | exosome component 10 | Affinity Capture-Western | BioGRID | 14527413 |
| EXOSC2 | RRP4 | Rrp4p | hRrp4p | p7 | exosome component 2 | Affinity Capture-Western | BioGRID | 14527413 |
| EXOSC4 | FLJ20591 | RRP41 | RRP41A | Rrp41p | SKI6 | Ski6p | hRrp41p | p12A | exosome component 4 | - | HPRD,BioGRID | 14527413 |
| LSM1 | CASM | YJL124C | LSM1 homolog, U6 small nuclear RNA associated (S. cerevisiae) | Two-hybrid | BioGRID | 15231747 |
| MAP1A | FLJ77111 | MAP1L | MTAP1A | microtubule-associated protein 1A | - | HPRD,BioGRID | 15231747 |
| PARN | DAN | poly(A)-specific ribonuclease (deadenylation nuclease) | Affinity Capture-Western | BioGRID | 14527413 |
| RPS15 | MGC111130 | RIG | ribosomal protein S15 | - | HPRD,BioGRID | 15231747 |
| RPS25 | - | ribosomal protein S25 | - | HPRD,BioGRID | 15231747 |
| SMG1 | 61E3.4 | ATX | KIAA0421 | LIP | PI-3-kinase-related kinase SMG-1 | Affinity Capture-Western | BioGRID | 11544179 |
| SRRM1 | 160-KD | MGC39488 | POP101 | SRM160 | serine/arginine repetitive matrix 1 | - | HPRD,BioGRID | 12093754 |
| UPF1 | FLJ43809 | FLJ46894 | HUPF1 | KIAA0221 | NORF1 | RENT1 | pNORF1 | UPF1 regulator of nonsense transcripts homolog (yeast) | Upf1 interacts with Upf2. | BIND | 15680326 |
| UPF1 | FLJ43809 | FLJ46894 | HUPF1 | KIAA0221 | NORF1 | RENT1 | pNORF1 | UPF1 regulator of nonsense transcripts homolog (yeast) | - | HPRD,BioGRID | 11073994 |
| UPF1 | FLJ43809 | FLJ46894 | HUPF1 | KIAA0221 | NORF1 | RENT1 | pNORF1 | UPF1 regulator of nonsense transcripts homolog (yeast) | - | HPRD | 11073994 |12723973 |
| UPF2 | DKFZp434D222 | HUPF2 | KIAA1408 | MGC138834 | MGC138835 | RENT2 | smg-3 | UPF2 regulator of nonsense transcripts homolog (yeast) | Affinity Capture-Western | BioGRID | 14527413 |
| UPF3A | HUPF3A | RENT3A | UPF3 | UPF3 regulator of nonsense transcripts homolog A (yeast) | - | HPRD,BioGRID | 11163187 |
| UPF3B | HUPF3B | MRXS14 | RENT3B | UPF3X | UPF3 regulator of nonsense transcripts homolog B (yeast) | - | HPRD,BioGRID | 15231747 |
| XRN1 | DKFZp434P0721 | DKFZp686B22225 | DKFZp686F19113 | FLJ41903 | SEP1 | 5'-3' exoribonuclease 1 | - | HPRD,BioGRID | 14527413 |
| XRN2 | - | 5'-3' exoribonuclease 2 | - | HPRD,BioGRID | 15231747 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | 176 | 57 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR UP | 156 | 101 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |