Gene Page: GFAP
Summary ?
| GeneID | 2670 |
| Symbol | GFAP |
| Synonyms | ALXDRD |
| Description | glial fibrillary acidic protein |
| Reference | MIM:137780|HGNC:HGNC:4235|Ensembl:ENSG00000131095|HPRD:00675|Vega:OTTHUMG00000179866 |
| Gene type | protein-coding |
| Map location | 17q21 |
| Pascal p-value | 0.359 |
| Sherlock p-value | 0.757 |
| Fetal beta | -1.865 |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0584 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
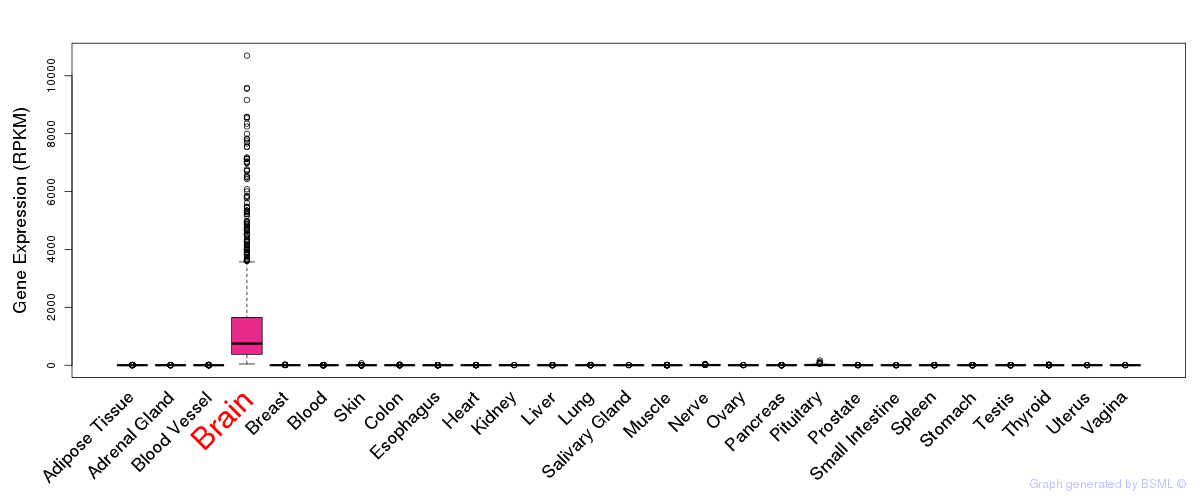
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| APBB3 | 0.88 | 0.87 |
| SGSM3 | 0.86 | 0.86 |
| ABCB9 | 0.85 | 0.83 |
| MAP3K12 | 0.85 | 0.83 |
| NICN1 | 0.85 | 0.82 |
| C16orf58 | 0.85 | 0.80 |
| KAT2A | 0.85 | 0.83 |
| TMEM25 | 0.84 | 0.84 |
| SCYL1 | 0.84 | 0.82 |
| PCYOX1L | 0.84 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.2 | -0.66 | -0.49 |
| AF347015.21 | -0.65 | -0.51 |
| AL139819.3 | -0.65 | -0.63 |
| AF347015.31 | -0.65 | -0.51 |
| AF347015.8 | -0.64 | -0.50 |
| MT-CO2 | -0.64 | -0.49 |
| MT-CYB | -0.63 | -0.49 |
| AF347015.18 | -0.62 | -0.53 |
| AF347015.26 | -0.62 | -0.47 |
| AF347015.15 | -0.60 | -0.45 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005200 | structural constituent of cytoskeleton | TAS | 2740350 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005882 | intermediate filament | TAS | 2740350 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CEP76 | C18orf9 | FLJ12542 | HsT1705 | centrosomal protein 76kDa | Two-hybrid | BioGRID | 16189514 |
| GRAP2 | GADS | GRAP-2 | GRB2L | GRBLG | GRID | GRPL | GrbX | Grf40 | Mona | P38 | GRB2-related adaptor protein 2 | Two-hybrid | BioGRID | 16189514 |
| KIAA0408 | FLJ43995 | RP3-403A15.2 | KIAA0408 | Two-hybrid | BioGRID | 16189514 |
| MEN1 | MEAI | SCG2 | multiple endocrine neoplasia I | - | HPRD,BioGRID | 12169273 |
| MYO15B | FLJ21606 | FLJ22686 | FLJ40577 | FLJ44494 | MYO15BP | myosin XVB pseudogene | Two-hybrid | BioGRID | 16189514 |
| PDLIM1 | CLIM1 | CLP-36 | CLP36 | hCLIM1 | PDZ and LIM domain 1 | Two-hybrid | BioGRID | 16189514 |
| PDLIM7 | LMP1 | PDZ and LIM domain 7 (enigma) | Two-hybrid | BioGRID | 16189514 |
| PDZK1 | CAP70 | CLAMP | NHERF3 | PDZD1 | PDZ domain containing 1 | Two-hybrid | BioGRID | 16189514 |
| POM121 | DKFZp586G1822 | DKFZp586P2220 | FLJ41820 | KIAA0618 | MGC3792 | POM121A | POM121 membrane glycoprotein (rat) | Two-hybrid | BioGRID | 16189514 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | - | HPRD,BioGRID | 12058025 |
| PSEN2 | AD3L | AD4 | PS2 | STM2 | presenilin 2 (Alzheimer disease 4) | Two-hybrid | BioGRID | 12058025 |
| RIBC2 | C22orf11 | RIB43A domain with coiled-coils 2 | Two-hybrid | BioGRID | 16189514 |
| S100A1 | S100 | S100-alpha | S100A | S100 calcium binding protein A1 | - | HPRD | 10510252 |
| S100B | NEF | S100 | S100beta | S100 calcium binding protein B | - | HPRD | 10510252 |
| SH3YL1 | DKFZp586F1318 | FLJ39121 | Ray | SH3 domain containing, Ysc84-like 1 (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD | 11379820 |
| TUBGCP4 | 76P | FLJ14797 | GCP4 | tubulin, gamma complex associated protein 4 | Two-hybrid | BioGRID | 16189514 |
| VIM | FLJ36605 | vimentin | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR SIGNALING BY ERBB4 | 38 | 30 | All SZGR 2.0 genes in this pathway |
| COLIN PILOCYTIC ASTROCYTOMA VS GLIOBLASTOMA UP | 35 | 32 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| CHEN LUNG CANCER SURVIVAL | 28 | 22 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR DN | 86 | 62 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX UP | 89 | 59 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| NIELSEN SCHWANNOMA UP | 18 | 16 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| LEIN ASTROCYTE MARKERS | 42 | 35 | All SZGR 2.0 genes in this pathway |
| LEIN PONS MARKERS | 89 | 59 | All SZGR 2.0 genes in this pathway |
| HOQUE METHYLATED IN CANCER | 56 | 45 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| CUI GLUCOSE DEPRIVATION | 60 | 44 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3 UNMETHYLATED | 228 | 119 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| DEMAGALHAES AGING UP | 55 | 39 | All SZGR 2.0 genes in this pathway |