Gene Page: ATP5S
Summary ?
| GeneID | 27109 |
| Symbol | ATP5S |
| Synonyms | ATPW|FB|HSU79253 |
| Description | ATP synthase, H+ transporting, mitochondrial Fo complex subunit s (factor B) |
| Reference | HGNC:HGNC:18799|Ensembl:ENSG00000125375|HPRD:12503|Vega:OTTHUMG00000170866 |
| Gene type | protein-coding |
| Map location | 14q21.3 |
| Pascal p-value | 0.795 |
| Sherlock p-value | 0.782 |
| Fetal beta | -0.185 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Frontal Cortex BA9 Nucleus accumbens basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15383574 | 14 | 50779169 | ATP5S | 1.37E-10 | -0.013 | 4.9E-7 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11157721 | chr14 | 50516713 | ATP5S | 27109 | 0.12 | cis | ||
| rs1955926 | chr14 | 50699867 | ATP5S | 27109 | 6.435E-5 | cis | ||
| rs8013289 | chr14 | 50727009 | ATP5S | 27109 | 2.496E-5 | cis | ||
| rs8013174 | chr14 | 50727108 | ATP5S | 27109 | 4.81E-5 | cis | ||
| rs4901008 | chr14 | 50737668 | ATP5S | 27109 | 1.045E-4 | cis | ||
| rs12432105 | chr14 | 50747336 | ATP5S | 27109 | 3.378E-5 | cis | ||
| rs12433794 | chr14 | 50786003 | ATP5S | 27109 | 2.904E-5 | cis | ||
| rs7154788 | chr14 | 50798036 | ATP5S | 27109 | 9.267E-5 | cis | ||
| rs7161563 | chr14 | 50799125 | ATP5S | 27109 | 3.26E-6 | cis | ||
| rs4901022 | chr14 | 50834258 | ATP5S | 27109 | 0.06 | cis | ||
| rs1955926 | chr14 | 50699867 | ATP5S | 27109 | 0.01 | trans | ||
| rs8013289 | chr14 | 50727009 | ATP5S | 27109 | 0 | trans | ||
| rs8013174 | chr14 | 50727108 | ATP5S | 27109 | 0.01 | trans | ||
| rs4901008 | chr14 | 50737668 | ATP5S | 27109 | 0.01 | trans | ||
| rs12432105 | chr14 | 50747336 | ATP5S | 27109 | 0 | trans | ||
| rs12433794 | chr14 | 50786003 | ATP5S | 27109 | 0 | trans | ||
| rs7154788 | chr14 | 50798036 | ATP5S | 27109 | 0.01 | trans | ||
| rs7161563 | chr14 | 50799125 | ATP5S | 27109 | 5.704E-4 | trans | ||
| rs8013289 | 14 | 50727010 | ATP5S | ENSG00000125375.10 | 8.583E-7 | 0.01 | -54299 | gtex_brain_ba24 |
| rs8013174 | 14 | 50727109 | ATP5S | ENSG00000125375.10 | 9.627E-7 | 0.01 | -54200 | gtex_brain_ba24 |
| rs4901005 | 14 | 50728736 | ATP5S | ENSG00000125375.10 | 9.627E-7 | 0.01 | -52573 | gtex_brain_ba24 |
| rs12431485 | 14 | 50734100 | ATP5S | ENSG00000125375.10 | 2.06E-6 | 0.01 | -47209 | gtex_brain_ba24 |
| rs11157734 | 14 | 50739485 | ATP5S | ENSG00000125375.10 | 2.453E-6 | 0.01 | -41824 | gtex_brain_ba24 |
| rs11849020 | 14 | 50740605 | ATP5S | ENSG00000125375.10 | 2.453E-6 | 0.01 | -40704 | gtex_brain_ba24 |
| rs11849059 | 14 | 50740700 | ATP5S | ENSG00000125375.10 | 2.285E-6 | 0.01 | -40609 | gtex_brain_ba24 |
| rs1465159 | 14 | 50743732 | ATP5S | ENSG00000125375.10 | 2.453E-6 | 0.01 | -37577 | gtex_brain_ba24 |
| rs59450505 | 14 | 50745123 | ATP5S | ENSG00000125375.10 | 2.453E-6 | 0.01 | -36186 | gtex_brain_ba24 |
| rs12432105 | 14 | 50747337 | ATP5S | ENSG00000125375.10 | 2.453E-6 | 0.01 | -33972 | gtex_brain_ba24 |
| rs12889265 | 14 | 50748853 | ATP5S | ENSG00000125375.10 | 2.46E-6 | 0.01 | -32456 | gtex_brain_ba24 |
| rs12432978 | 14 | 50750316 | ATP5S | ENSG00000125375.10 | 2.453E-6 | 0.01 | -30993 | gtex_brain_ba24 |
| rs6572655 | 14 | 50751565 | ATP5S | ENSG00000125375.10 | 2.453E-6 | 0.01 | -29744 | gtex_brain_ba24 |
| rs11849603 | 14 | 50753242 | ATP5S | ENSG00000125375.10 | 2.453E-6 | 0.01 | -28067 | gtex_brain_ba24 |
| rs7154966 | 14 | 50753992 | ATP5S | ENSG00000125375.10 | 2.453E-6 | 0.01 | -27317 | gtex_brain_ba24 |
| rs12881896 | 14 | 50755991 | ATP5S | ENSG00000125375.10 | 2.453E-6 | 0.01 | -25318 | gtex_brain_ba24 |
| rs9888567 | 14 | 50757131 | ATP5S | ENSG00000125375.10 | 2.453E-6 | 0.01 | -24178 | gtex_brain_ba24 |
| rs7142135 | 14 | 50759678 | ATP5S | ENSG00000125375.10 | 2.452E-6 | 0.01 | -21631 | gtex_brain_ba24 |
| rs7144289 | 14 | 50761847 | ATP5S | ENSG00000125375.10 | 2.177E-6 | 0.01 | -19462 | gtex_brain_ba24 |
| rs4901011 | 14 | 50768483 | ATP5S | ENSG00000125375.10 | 2.453E-6 | 0.01 | -12826 | gtex_brain_ba24 |
| rs2297993 | 14 | 50769327 | ATP5S | ENSG00000125375.10 | 2.453E-6 | 0.01 | -11982 | gtex_brain_ba24 |
| rs2297994 | 14 | 50769455 | ATP5S | ENSG00000125375.10 | 2.453E-6 | 0.01 | -11854 | gtex_brain_ba24 |
| rs2297995 | 14 | 50769717 | ATP5S | ENSG00000125375.10 | 2.453E-6 | 0.01 | -11592 | gtex_brain_ba24 |
| rs1974327 | 14 | 50769901 | ATP5S | ENSG00000125375.10 | 2.453E-6 | 0.01 | -11408 | gtex_brain_ba24 |
| rs8018446 | 14 | 50772403 | ATP5S | ENSG00000125375.10 | 2.406E-6 | 0.01 | -8906 | gtex_brain_ba24 |
| rs12880323 | 14 | 50773576 | ATP5S | ENSG00000125375.10 | 2.584E-6 | 0.01 | -7733 | gtex_brain_ba24 |
| rs6572656 | 14 | 50775319 | ATP5S | ENSG00000125375.10 | 2.482E-6 | 0.01 | -5990 | gtex_brain_ba24 |
| rs1967919 | 14 | 50786841 | ATP5S | ENSG00000125375.10 | 2.458E-7 | 0.01 | 5532 | gtex_brain_ba24 |
| rs7140879 | 14 | 50799643 | ATP5S | ENSG00000125375.10 | 2.724E-6 | 0.01 | 18334 | gtex_brain_ba24 |
| rs200027456 | 14 | 50800242 | ATP5S | ENSG00000125375.10 | 3.105E-6 | 0.01 | 18933 | gtex_brain_ba24 |
| rs4901017 | 14 | 50800618 | ATP5S | ENSG00000125375.10 | 1.858E-6 | 0.01 | 19309 | gtex_brain_ba24 |
Section II. Transcriptome annotation
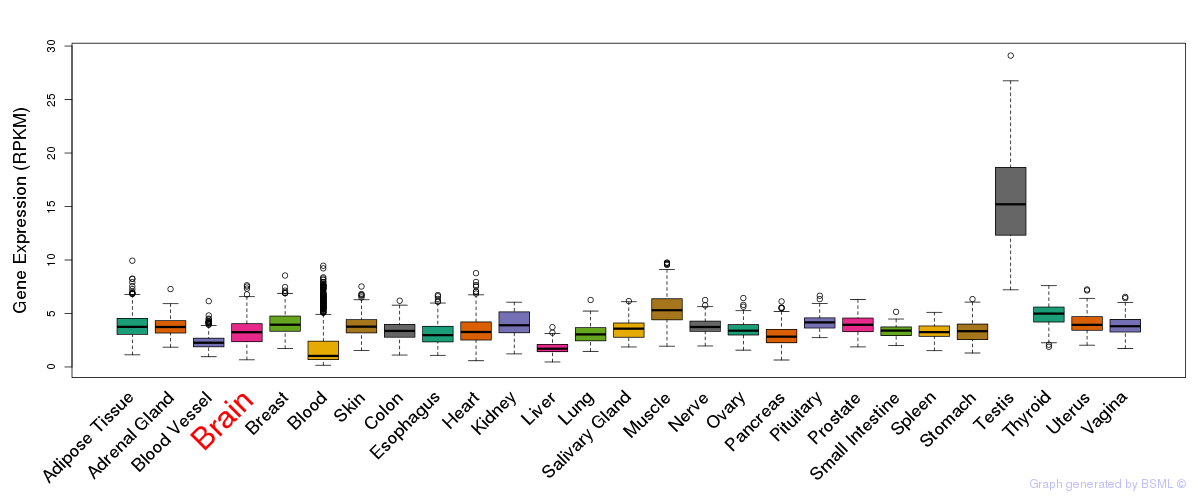
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIU SOX4 TARGETS UP | 137 | 94 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION UP | 119 | 66 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION MUSCLE DN | 51 | 28 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| LIN NPAS4 TARGETS UP | 163 | 100 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |