Gene Page: TOR1B
Summary ?
| GeneID | 27348 |
| Symbol | TOR1B |
| Synonyms | DQ1 |
| Description | torsin family 1 member B |
| Reference | MIM:608050|HGNC:HGNC:11995|Ensembl:ENSG00000136816|HPRD:16270|Vega:OTTHUMG00000020795 |
| Gene type | protein-coding |
| Map location | 9q34 |
| Pascal p-value | 0.015 |
| Fetal beta | 0.099 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Putamen basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs13290805 | 9 | 132570924 | TOR1B | ENSG00000136816.11 | 1.207E-6 | 0.01 | 5492 | gtex_brain_putamen_basal |
| rs12634 | 9 | 132573536 | TOR1B | ENSG00000136816.11 | 1.148E-6 | 0.01 | 8104 | gtex_brain_putamen_basal |
| rs10642255 | 9 | 132582425 | TOR1B | ENSG00000136816.11 | 5.854E-7 | 0.01 | 16993 | gtex_brain_putamen_basal |
| rs34429226 | 9 | 132583929 | TOR1B | ENSG00000136816.11 | 1.003E-6 | 0.01 | 18497 | gtex_brain_putamen_basal |
| rs2296793 | 9 | 132585058 | TOR1B | ENSG00000136816.11 | 6.217E-7 | 0.01 | 19626 | gtex_brain_putamen_basal |
| rs8113 | 9 | 132589606 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 24174 | gtex_brain_putamen_basal |
| rs8719 | 9 | 132589633 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 24201 | gtex_brain_putamen_basal |
| rs1806988 | 9 | 132590352 | TOR1B | ENSG00000136816.11 | 1.074E-6 | 0.01 | 24920 | gtex_brain_putamen_basal |
| rs1806989 | 9 | 132590353 | TOR1B | ENSG00000136816.11 | 1.089E-6 | 0.01 | 24921 | gtex_brain_putamen_basal |
| rs732074 | 9 | 132590431 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 24999 | gtex_brain_putamen_basal |
| rs35168956 | 9 | 132590837 | TOR1B | ENSG00000136816.11 | 8.572E-7 | 0.01 | 25405 | gtex_brain_putamen_basal |
| rs45441205 | 9 | 132590843 | TOR1B | ENSG00000136816.11 | 8.572E-7 | 0.01 | 25411 | gtex_brain_putamen_basal |
| rs45562032 | 9 | 132590860 | TOR1B | ENSG00000136816.11 | 3.536E-7 | 0.01 | 25428 | gtex_brain_putamen_basal |
| rs45581233 | 9 | 132590912 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 25480 | gtex_brain_putamen_basal |
| rs35075313 | 9 | 132590997 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 25565 | gtex_brain_putamen_basal |
| rs55962905 | 9 | 132591272 | TOR1B | ENSG00000136816.11 | 9.591E-7 | 0.01 | 25840 | gtex_brain_putamen_basal |
| rs7032945 | 9 | 132591372 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 25940 | gtex_brain_putamen_basal |
| rs3818553 | 9 | 132591509 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 26077 | gtex_brain_putamen_basal |
| rs41278732 | 9 | 132591992 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 26560 | gtex_brain_putamen_basal |
| rs13290140 | 9 | 132592211 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 26779 | gtex_brain_putamen_basal |
| rs34908998 | 9 | 132592636 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 27204 | gtex_brain_putamen_basal |
| rs34560496 | 9 | 132592663 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 27231 | gtex_brain_putamen_basal |
| rs13286552 | 9 | 132592775 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 27343 | gtex_brain_putamen_basal |
| rs2062801 | 9 | 132593982 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 28550 | gtex_brain_putamen_basal |
| rs2062800 | 9 | 132594362 | TOR1B | ENSG00000136816.11 | 1.891E-6 | 0.01 | 28930 | gtex_brain_putamen_basal |
| rs2062799 | 9 | 132594378 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 28946 | gtex_brain_putamen_basal |
| rs58979947 | 9 | 132594658 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 29226 | gtex_brain_putamen_basal |
| rs2062798 | 9 | 132594718 | TOR1B | ENSG00000136816.11 | 8.886E-7 | 0.01 | 29286 | gtex_brain_putamen_basal |
| rs34862553 | 9 | 132594938 | TOR1B | ENSG00000136816.11 | 1.983E-6 | 0.01 | 29506 | gtex_brain_putamen_basal |
| rs34904094 | 9 | 132594984 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 29552 | gtex_brain_putamen_basal |
| rs72755238 | 9 | 132595664 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 30232 | gtex_brain_putamen_basal |
| rs45507500 | 9 | 132595685 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 30253 | gtex_brain_putamen_basal |
| rs2274508 | 9 | 132596071 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 30639 | gtex_brain_putamen_basal |
| rs67061074 | 9 | 132596159 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 30727 | gtex_brain_putamen_basal |
| rs3814547 | 9 | 132597021 | TOR1B | ENSG00000136816.11 | 1.906E-6 | 0.01 | 31589 | gtex_brain_putamen_basal |
| rs11790613 | 9 | 132597621 | TOR1B | ENSG00000136816.11 | 1.894E-6 | 0.01 | 32189 | gtex_brain_putamen_basal |
| rs13294710 | 9 | 132597721 | TOR1B | ENSG00000136816.11 | 1.512E-6 | 0.01 | 32289 | gtex_brain_putamen_basal |
| rs7037982 | 9 | 132602208 | TOR1B | ENSG00000136816.11 | 2.173E-6 | 0.01 | 36776 | gtex_brain_putamen_basal |
| rs12342350 | 9 | 132603071 | TOR1B | ENSG00000136816.11 | 1.225E-6 | 0.01 | 37639 | gtex_brain_putamen_basal |
| rs13297359 | 9 | 132603396 | TOR1B | ENSG00000136816.11 | 1.124E-6 | 0.01 | 37964 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
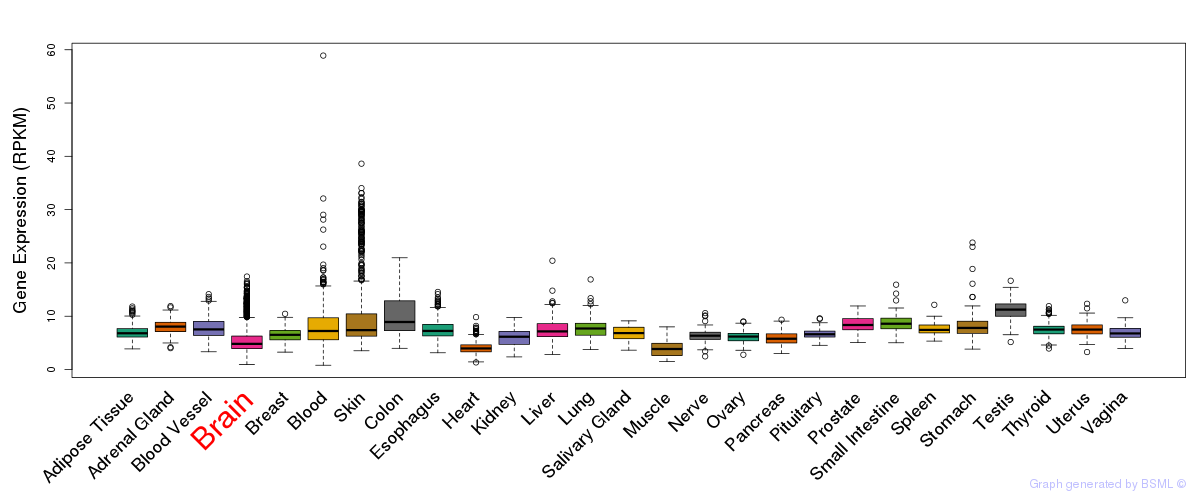
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER WITH LOH IN CHR9Q | 116 | 71 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| PETRETTO HEART MASS QTL CIS UP | 28 | 15 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS DN | 314 | 188 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |