Gene Page: SFN
Summary ?
| GeneID | 2810 |
| Symbol | SFN |
| Synonyms | YWHAS |
| Description | stratifin |
| Reference | MIM:601290|HGNC:HGNC:10773|Ensembl:ENSG00000175793|HPRD:03185|Vega:OTTHUMG00000004093 |
| Gene type | protein-coding |
| Map location | 1p36.11 |
| Pascal p-value | 0.01 |
| Fetal beta | 0.237 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08366132 | 1 | 27191899 | SFN | 3.2E-8 | -0.008 | 9.67E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6703905 | chr1 | 89656748 | SFN | 2810 | 0 | trans | ||
| rs7514271 | 0 | SFN | 2810 | 0.02 | trans | |||
| snp_a-1933592 | 0 | SFN | 2810 | 0 | trans | |||
| rs1347659 | chr2 | 142478063 | SFN | 2810 | 0.04 | trans | ||
| rs10497429 | chr2 | 175600890 | SFN | 2810 | 0 | trans | ||
| rs10513763 | chr3 | 179386743 | SFN | 2810 | 0.01 | trans | ||
| rs16830710 | chr3 | 179405727 | SFN | 2810 | 0.01 | trans | ||
| rs16896993 | chr4 | 18422329 | SFN | 2810 | 0.1 | trans | ||
| rs10518332 | chr4 | 120395972 | SFN | 2810 | 0.11 | trans | ||
| rs17359910 | chr4 | 120570768 | SFN | 2810 | 0.05 | trans | ||
| rs7737662 | chr5 | 52624188 | SFN | 2810 | 0 | trans | ||
| rs11134564 | chr5 | 168688649 | SFN | 2810 | 0.03 | trans | ||
| rs226946 | chr6 | 3717139 | SFN | 2810 | 0.01 | trans | ||
| rs226945 | chr6 | 3717178 | SFN | 2810 | 0.01 | trans | ||
| rs226966 | chr6 | 3720300 | SFN | 2810 | 0 | trans | ||
| rs715207 | chr6 | 88370956 | SFN | 2810 | 0.16 | trans | ||
| rs16883361 | chr8 | 113295049 | SFN | 2810 | 0.02 | trans | ||
| rs10970106 | chr9 | 31107128 | SFN | 2810 | 0.01 | trans | ||
| rs1475793 | chr9 | 31107450 | SFN | 2810 | 0.02 | trans | ||
| rs10983341 | chr9 | 119530424 | SFN | 2810 | 0.14 | trans | ||
| rs4750265 | chr10 | 12839958 | SFN | 2810 | 0.01 | trans | ||
| rs2482031 | chr10 | 12841159 | SFN | 2810 | 0.01 | trans | ||
| rs360133 | 0 | SFN | 2810 | 0.04 | trans | |||
| rs11823069 | chr11 | 10639788 | SFN | 2810 | 0.07 | trans | ||
| rs10500729 | chr11 | 10644598 | SFN | 2810 | 0.07 | trans | ||
| rs16908086 | chr11 | 10646521 | SFN | 2810 | 0.07 | trans | ||
| rs10500730 | chr11 | 10646550 | SFN | 2810 | 0.07 | trans | ||
| rs16908093 | chr11 | 10646630 | SFN | 2810 | 0.07 | trans | ||
| rs11833888 | chr12 | 59350152 | SFN | 2810 | 0.09 | trans | ||
| rs2993290 | chr13 | 113647189 | SFN | 2810 | 0 | trans | ||
| rs7152458 | chr14 | 96889899 | SFN | 2810 | 0.13 | trans | ||
| rs16944395 | chr15 | 61897800 | SFN | 2810 | 0.01 | trans | ||
| rs7193958 | chr16 | 34293326 | SFN | 2810 | 0.07 | trans | ||
| rs16946662 | chr16 | 48976991 | SFN | 2810 | 0.05 | trans | ||
| snp_a-1812923 | 0 | SFN | 2810 | 0.05 | trans | |||
| rs8060581 | chr16 | 80192604 | SFN | 2810 | 0.08 | trans | ||
| rs3803656 | chr16 | 81551505 | SFN | 2810 | 0.04 | trans | ||
| rs9892858 | chr17 | 60844131 | SFN | 2810 | 0.09 | trans | ||
| rs7208997 | chr17 | 60846657 | SFN | 2810 | 0.09 | trans |
Section II. Transcriptome annotation
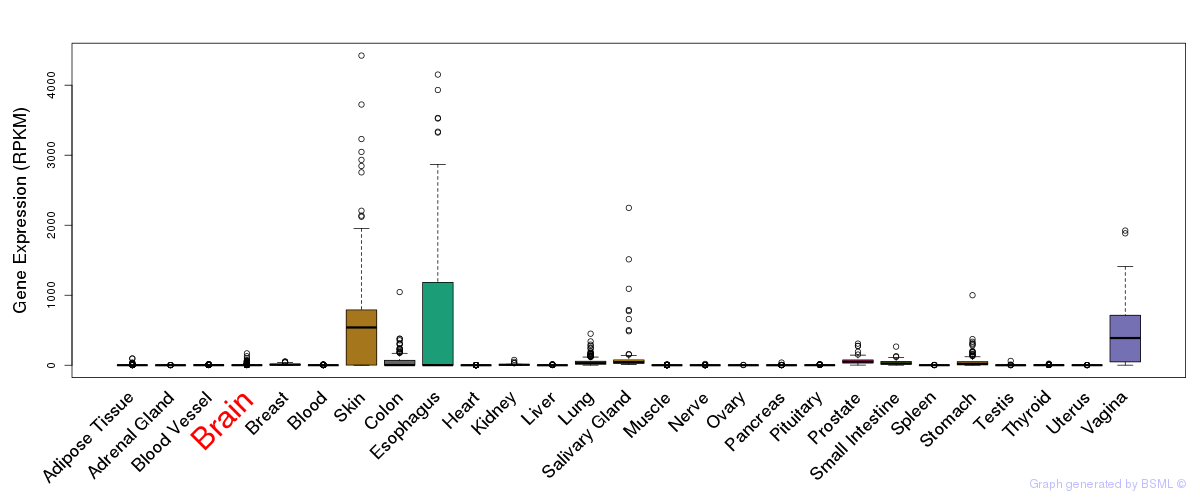
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CNTNAP1 | 0.92 | 0.89 |
| KCNJ9 | 0.90 | 0.83 |
| DNM1 | 0.90 | 0.81 |
| PTPRN | 0.89 | 0.82 |
| TMEM38A | 0.89 | 0.91 |
| HTR5A | 0.89 | 0.82 |
| SLC12A5 | 0.89 | 0.81 |
| SYNGR1 | 0.89 | 0.76 |
| MFSD4 | 0.89 | 0.86 |
| ZFYVE28 | 0.89 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GTF3C6 | -0.55 | -0.58 |
| FAM36A | -0.52 | -0.52 |
| C9orf46 | -0.52 | -0.62 |
| RBMX2 | -0.52 | -0.67 |
| BCL7C | -0.52 | -0.63 |
| TUBB2B | -0.51 | -0.60 |
| KIAA1949 | -0.51 | -0.48 |
| TRAF4 | -0.49 | -0.57 |
| YBX1 | -0.49 | -0.51 |
| DYNLT1 | -0.49 | -0.66 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABLIM1 | ABLIM | DKFZp781D0148 | FLJ14564 | KIAA0059 | LIMAB1 | LIMATIN | MGC1224 | actin binding LIM protein 1 | Affinity Capture-MS | BioGRID | 15778465 |
| ANKS1A | ANKS1 | KIAA0229 | MGC42354 | ankyrin repeat and sterile alpha motif domain containing 1A | Affinity Capture-MS | BioGRID | 15778465 |
| APC | BTPS2 | DP2 | DP2.5 | DP3 | GS | adenomatous polyposis coli | Affinity Capture-MS | BioGRID | 15778465 |
| APLP2 | APPH | APPL2 | CDEBP | amyloid beta (A4) precursor-like protein 2 | Affinity Capture-MS | BioGRID | 15778465 |
| ARAF | A-RAF | ARAF1 | PKS2 | RAFA1 | v-raf murine sarcoma 3611 viral oncogene homolog | Affinity Capture-MS | BioGRID | 15778465 |
| ARHGAP11A | KIAA0013 | MGC70740 | Rho GTPase activating protein 11A | Affinity Capture-MS | BioGRID | 15778465 |
| ARHGAP21 | ARHGAP10 | DKFZp761L0424 | FLJ33323 | FLJ90108 | Rho GTPase activating protein 21 | Affinity Capture-MS | BioGRID | 15778465 |
| ARHGEF16 | GEF16 | NBR | Rho guanine exchange factor (GEF) 16 | Affinity Capture-MS | BioGRID | 15778465 |
| ARHGEF17 | FLJ90019 | KIAA0337 | P164RHOGEF | TEM4 | p164-RhoGEF | Rho guanine nucleotide exchange factor (GEF) 17 | Affinity Capture-MS | BioGRID | 15778465 |
| ARHGEF5 | DKFZp686N1969 | GEF5 | P60 | TIM | TIM1 | Rho guanine nucleotide exchange factor (GEF) 5 | Affinity Capture-MS | BioGRID | 15778465 |
| BAD | BBC2 | BCL2L8 | BCL2-associated agonist of cell death | Two-hybrid | BioGRID | 16189514 |
| BAX | BCL2L4 | BCL2-associated X protein | - | HPRD | 11574543 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | Affinity Capture-MS | BioGRID | 15778465 |
| BRAF | B-RAF1 | BRAF1 | FLJ95109 | MGC126806 | MGC138284 | RAFB1 | v-raf murine sarcoma viral oncogene homolog B1 | Affinity Capture-MS | BioGRID | 15778465 |
| C9orf61 | MGC142243 | MGC142245 | RP11-548B3.1 | X123 | chromosome 9 open reading frame 61 | Two-hybrid | BioGRID | 16189514 |
| CCAR1 | CARP-1 | CARP1 | MGC44628 | RP11-437A18.1 | cell division cycle and apoptosis regulator 1 | - | HPRD | 12816952 |
| CDC2 | CDC28A | CDK1 | DKFZp686L20222 | MGC111195 | cell division cycle 2, G1 to S and G2 to M | - | HPRD,BioGRID | 10524633 |
| CGN | DKFZp779N1112 | FLJ39281 | KIAA1319 | cingulin | Affinity Capture-MS | BioGRID | 15778465 |
| CHEK1 | CHK1 | CHK1 checkpoint homolog (S. pombe) | Affinity Capture-Western | BioGRID | 11896572 |
| CHST1 | C6ST | KS6ST | KSGal6ST | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | Two-hybrid | BioGRID | 16189514 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | Affinity Capture-MS | BioGRID | 15778465 |
| CYTH2 | ARNO | CTS18 | CTS18.1 | PSCD2 | PSCD2L | SEC7L | Sec7p-L | Sec7p-like | cytohesin 2 | Affinity Capture-MS | BioGRID | 15778465 |
| DTX2 | KIAA1528 | MGC71098 | RNF58 | deltex homolog 2 (Drosophila) | Affinity Capture-MS | BioGRID | 15778465 |
| DYRK1A | DYRK | DYRK1 | HP86 | MNB | MNBH | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | Affinity Capture-MS | BioGRID | 15778465 |
| ERRFI1 | GENE-33 | MIG-6 | MIG6 | RALT | ERBB receptor feedback inhibitor 1 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 15778465 |
| FUSIP1 | FUSIP2 | NSSR | SFRS13 | SRp38 | SRrp40 | TASR | TASR1 | TASR2 | FUS interacting protein (serine/arginine-rich) 1 | Affinity Capture-MS | BioGRID | 15778465 |
| GAN | FLJ38059 | GAN1 | KLHL16 | gigaxonin | Affinity Capture-MS | BioGRID | 15778465 |
| GPRIN2 | GRIN2 | KIAA0514 | MGC15171 | G protein regulated inducer of neurite outgrowth 2 | Two-hybrid | BioGRID | 16189514 |
| GRB7 | - | growth factor receptor-bound protein 7 | Affinity Capture-MS | BioGRID | 15778465 |
| HDAC4 | HA6116 | HD4 | HDAC-A | HDACA | KIAA0288 | histone deacetylase 4 | Affinity Capture-MS | BioGRID | 15778465 |
| HDAC5 | FLJ90614 | HD5 | NY-CO-9 | histone deacetylase 5 | - | HPRD | 15367659 |
| HDAC7 | DKFZp586J0917 | FLJ99588 | HD7A | HDAC7A | histone deacetylase 7 | Affinity Capture-MS | BioGRID | 15778465 |
| HNRNPU | HNRPU | SAF-A | U21.1 | heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) | Affinity Capture-MS | BioGRID | 15778465 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | Affinity Capture-MS | BioGRID | 15778465 |
| IRS2 | - | insulin receptor substrate 2 | Affinity Capture-MS | BioGRID | 15778465 |
| ISCU | 2310020H20Rik | HML | ISU2 | MGC74517 | NIFU | NIFUN | hnifU | iron-sulfur cluster scaffold homolog (E. coli) | Affinity Capture-MS | BioGRID | 15778465 |
| ITCH | AIF4 | AIP4 | NAPP1 | dJ468O1.1 | itchy E3 ubiquitin protein ligase homolog (mouse) | Affinity Capture-MS | BioGRID | 15778465 |
| JUB | Ajuba | MGC15563 | jub, ajuba homolog (Xenopus laevis) | Affinity Capture-MS Affinity Capture-Western | BioGRID | 15778465 |
| KIAA0408 | FLJ43995 | RP3-403A15.2 | KIAA0408 | Two-hybrid | BioGRID | 16189514 |
| KIF23 | CHO1 | KNSL5 | MKLP-1 | MKLP1 | kinesin family member 23 | Affinity Capture-MS | BioGRID | 15778465 |
| KIF5B | KINH | KNS | KNS1 | UKHC | kinesin family member 5B | Affinity Capture-MS | BioGRID | 15778465 |
| KLC1 | KLC | KNS2 | KNS2A | MGC15245 | kinesin light chain 1 | Affinity Capture-MS | BioGRID | 15778465 |
| KLC2 | FLJ12387 | kinesin light chain 2 | Affinity Capture-MS | BioGRID | 15778465 |
| KLHDC2 | HCLP-1 | LCP | kelch domain containing 2 | Affinity Capture-MS | BioGRID | 15778465 |
| KRT18 | CYK18 | K18 | keratin 18 | - | HPRD | 11917136 |
| LAD1 | LadA | MGC10355 | ladinin 1 | Affinity Capture-MS | BioGRID | 15778465 |
| LMO7 | FBX20 | FBXO20 | KIAA0858 | LOMP | LIM domain 7 | Affinity Capture-MS | BioGRID | 15778465 |
| LSR | LISCH7 | MGC10659 | MGC48312 | MGC48503 | lipolysis stimulated lipoprotein receptor | Affinity Capture-MS | BioGRID | 15778465 |
| MAGI1 | AIP3 | BAIAP1 | BAP1 | MAGI-1 | TNRC19 | WWP3 | membrane associated guanylate kinase, WW and PDZ domain containing 1 | Affinity Capture-MS | BioGRID | 15778465 |
| MAP3K2 | MEKK2 | MEKK2B | mitogen-activated protein kinase kinase kinase 2 | Affinity Capture-MS | BioGRID | 15778465 |
| MAPKAP1 | JC310 | MGC2745 | MIP1 | SIN1 | SIN1b | SIN1g | mitogen-activated protein kinase associated protein 1 | Affinity Capture-MS | BioGRID | 15778465 |
| MARK1 | KIAA1477 | MARK | MGC126512 | MGC126513 | MAP/microtubule affinity-regulating kinase 1 | Affinity Capture-MS | BioGRID | 15778465 |
| MARK2 | EMK1 | MGC99619 | PAR-1 | Par1b | MAP/microtubule affinity-regulating kinase 2 | Affinity Capture-MS | BioGRID | 15778465 |
| MARK3 | CTAK1 | KP78 | PAR1A | MAP/microtubule affinity-regulating kinase 3 | Affinity Capture-MS Two-hybrid | BioGRID | 15778465 |16189514 |
| MPRIP | KIAA0864 | M-RIP | RHOIP3 | p116Rip | myosin phosphatase Rho interacting protein | Affinity Capture-MS | BioGRID | 15778465 |
| MYCBP2 | DKFZp686M08244 | FLJ10106 | FLJ13826 | FLJ21597 | FLJ21646 | KIAA0916 | PAM | MYC binding protein 2 | Affinity Capture-MS | BioGRID | 15778465 |
| NEDD4L | FLJ33870 | KIAA0439 | NEDD4-2 | RSP5 | hNedd4-2 | neural precursor cell expressed, developmentally down-regulated 4-like | Affinity Capture-MS | BioGRID | 15778465 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD,BioGRID | 12730237 |
| OSBPL3 | DKFZp667P1518 | KIAA0704 | MGC21526 | ORP-3 | ORP3 | OSBP3 | oxysterol binding protein-like 3 | Affinity Capture-MS | BioGRID | 15778465 |
| PAK4 | - | p21 protein (Cdc42/Rac)-activated kinase 4 | Affinity Capture-MS | BioGRID | 15778465 |
| PARD3 | ASIP | Baz | Bazooka | FLJ21015 | PAR3 | PAR3alpha | PARD3A | SE2-5L16 | SE2-5LT1 | SE2-5T2 | par-3 partitioning defective 3 homolog (C. elegans) | Affinity Capture-MS | BioGRID | 15778465 |
| PARD3B | ALS2CR19 | MGC16131 | PAR3B | PAR3L | PAR3LC | PAR3beta | Par3Lb | par-3 partitioning defective 3 homolog B (C. elegans) | Affinity Capture-MS | BioGRID | 15778465 |
| PI4KB | PI4K-BETA | PI4KIIIbeta | PI4Kbeta | PIK4CB | pi4K92 | phosphatidylinositol 4-kinase, catalytic, beta | Affinity Capture-MS | BioGRID | 15778465 |
| PIK3C2B | C2-PI3K | DKFZp686G16234 | phosphoinositide-3-kinase, class 2, beta polypeptide | Affinity Capture-MS | BioGRID | 15778465 |
| PKP2 | ARVD9 | plakophilin 2 | Affinity Capture-MS | BioGRID | 15778465 |
| PKP3 | - | plakophilin 3 | Affinity Capture-MS | BioGRID | 15778465 |
| PLEKHF2 | FLJ13187 | PHAFIN2 | ZFYVE18 | pleckstrin homology domain containing, family F (with FYVE domain) member 2 | Two-hybrid | BioGRID | 16189514 |
| PLK4 | SAK | STK18 | polo-like kinase 4 (Drosophila) | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| PPFIBP1 | L2 | hSGT2 | hSgt2p | PTPRF interacting protein, binding protein 1 (liprin beta 1) | Affinity Capture-MS | BioGRID | 15778465 |
| PPFIBP2 | Cclp1 | DKFZp781K06126 | MGC42541 | PTPRF interacting protein, binding protein 2 (liprin beta 2) | Affinity Capture-MS | BioGRID | 15778465 |
| PPP1R3D | DKFZp781L2441 | PPP1R6 | protein phosphatase 1, regulatory (inhibitor) subunit 3D | Affinity Capture-MS | BioGRID | 15778465 |
| PTOV1 | ACID2 | DKFZp586I111 | MGC71475 | prostate tumor overexpressed 1 | Affinity Capture-MS | BioGRID | 15778465 |
| PTPN2 | PTPT | TC-PTP | TCELLPTP | TCPTP | protein tyrosine phosphatase, non-receptor type 2 | Affinity Capture-MS | BioGRID | 15778465 |
| PTPN3 | DKFZp686N0569 | PTPH1 | protein tyrosine phosphatase, non-receptor type 3 | Affinity Capture-MS | BioGRID | 15778465 |
| RACGAP1 | HsCYK-4 | ID-GAP | MgcRacGAP | Rac GTPase activating protein 1 | Affinity Capture-MS | BioGRID | 15778465 |
| RAE1 | FLJ30608 | MGC117333 | MGC126076 | MGC126077 | MIG14 | MRNP41 | Mnrp41 | dJ481F12.3 | dJ800J21.1 | RAE1 RNA export 1 homolog (S. pombe) | Affinity Capture-MS | BioGRID | 15778465 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | Affinity Capture-MS | BioGRID | 15778465 |
| RALGPS2 | FLJ10244 | FLJ25604 | KIAA0351 | dJ595C2.1 | Ral GEF with PH domain and SH3 binding motif 2 | Affinity Capture-MS | BioGRID | 15778465 |
| RHPN2 | RhoBP | p76RBE | rhophilin, Rho GTPase binding protein 2 | Affinity Capture-MS | BioGRID | 15778465 |
| RICS | GC-GAP | GRIT | KIAA0712 | MGC1892 | p200RhoGAP | p250GAP | Rho GTPase-activating protein | Affinity Capture-MS | BioGRID | 15778465 |
| RND3 | ARHE | Rho8 | RhoE | memB | Rho family GTPase 3 | Affinity Capture-MS | BioGRID | 15778465 |
| SASH1 | KIAA0790 | RP3-323M4.1 | SH3D6A | dJ323M4 | dJ323M4.1 | SAM and SH3 domain containing 1 | Affinity Capture-MS | BioGRID | 15778465 |
| SAV1 | SAV | WW45 | WWP4 | salvador homolog 1 (Drosophila) | Affinity Capture-MS | BioGRID | 15778465 |
| SH2D3A | NSP1 | SH2 domain containing 3A | Affinity Capture-MS | BioGRID | 15778465 |
| SH3BP4 | BOG25 | TTP | SH3-domain binding protein 4 | Affinity Capture-MS | BioGRID | 15778465 |
| SHCBP1 | FLJ22009 | MGC26900 | PAL | SHC SH2-domain binding protein 1 | Affinity Capture-MS | BioGRID | 15778465 |
| SHROOM3 | APXL3 | KIAA1481 | MSTP013 | SHRM | ShrmL | shroom family member 3 | Affinity Capture-MS | BioGRID | 15778465 |
| SIPA1L1 | DKFZp686G1344 | E6TP1 | KIAA0440 | signal-induced proliferation-associated 1 like 1 | Affinity Capture-MS | BioGRID | 15778465 |
| SIPA1L3 | SPAL3 | signal-induced proliferation-associated 1 like 3 | Affinity Capture-MS | BioGRID | 15778465 |
| SORBS2 | ARGBP2 | FLJ93447 | KIAA0777 | PRO0618 | sorbin and SH3 domain containing 2 | Affinity Capture-MS | BioGRID | 15778465 |
| SPIRE1 | MGC150621 | MGC150622 | Spir-1 | spire homolog 1 (Drosophila) | Affinity Capture-MS | BioGRID | 15778465 |
| SRRM2 | 300-KD | CWF21 | DKFZp686O15166 | FLJ21926 | FLJ22250 | KIAA0324 | MGC40295 | SRL300 | SRm300 | serine/arginine repetitive matrix 2 | Affinity Capture-MS | BioGRID | 15778465 |
| ST5 | DENND2B | HTS1 | MGC33090 | p126 | suppression of tumorigenicity 5 | Affinity Capture-MS | BioGRID | 15778465 |
| SYNJ2 | INPP5H | KIAA0348 | MGC44422 | synaptojanin 2 | Affinity Capture-MS | BioGRID | 15778465 |
| TBL3 | SAZD | transducin (beta)-like 3 | Two-hybrid | BioGRID | 16189514 |
| TJP2 | MGC26306 | X104 | ZO-2 | ZO2 | tight junction protein 2 (zona occludens 2) | Affinity Capture-MS | BioGRID | 15778465 |
| TNK1 | MGC46193 | tyrosine kinase, non-receptor, 1 | Affinity Capture-MS | BioGRID | 15778465 |
| TNS4 | CTEN | FLJ14950 | PP14434 | tensin 4 | Affinity Capture-MS | BioGRID | 15778465 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Affinity Capture-Western | BioGRID | 11896572 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with the SFN (14-3-3-sigma) promoter. | BIND | 15574328 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with 14-3-3-sigma promoter. | BIND | 16009130 |
| TRIM25 | EFP | RNF147 | Z147 | ZNF147 | tripartite motif-containing 25 | - | HPRD,BioGRID | 12075357 |
| TRIM32 | BBS11 | HT2A | LGMD2H | TATIP | tripartite motif-containing 32 | Affinity Capture-MS | BioGRID | 15778465 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | - | HPRD | 12438239 |
| USP8 | FLJ34456 | HumORF8 | KIAA0055 | MGC129718 | UBPY | ubiquitin specific peptidase 8 | Affinity Capture-MS | BioGRID | 15778465 |
| WDR68 | AN11 | HAN11 | WD repeat domain 68 | Affinity Capture-MS | BioGRID | 15778465 |
| WDYHV1 | C8orf32 | FLJ10204 | WDYHV motif containing 1 | Two-hybrid | BioGRID | 16189514 |
| WEE1 | DKFZp686I18166 | FLJ16446 | WEE1A | WEE1hu | WEE1 homolog (S. pombe) | Affinity Capture-MS | BioGRID | 15778465 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | Affinity Capture-MS | BioGRID | 15778465 |
| ZAK | AZK | MLK7 | MLT | MLTK | MRK | mlklak | sterile alpha motif and leucine zipper containing kinase AZK | Affinity Capture-MS | BioGRID | 15778465 |
| ZFP36 | G0S24 | GOS24 | NUP475 | RNF162A | TIS11 | TTP | zinc finger protein 36, C3H type, homolog (mouse) | - | HPRD | 11886850 |
| ZNF638 | DKFZp686P1231 | MGC26130 | MGC90196 | NP220 | ZFML | Zfp638 | zinc finger protein 638 | Affinity Capture-MS | BioGRID | 15778465 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG ALDOSTERONE REGULATED SODIUM REABSORPTION | 42 | 29 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | 51 | 41 | All SZGR 2.0 genes in this pathway |
| ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| PID P73PATHWAY | 79 | 59 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID FOXO PATHWAY | 49 | 43 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID P38 MK2 PATHWAY | 21 | 19 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID DELTA NP63 PATHWAY | 47 | 34 | All SZGR 2.0 genes in this pathway |
| PID A6B1 A6B4 INTEGRIN PATHWAY | 46 | 35 | All SZGR 2.0 genes in this pathway |
| PID INSULIN GLUCOSE PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI AKT PATHWAY | 35 | 30 | All SZGR 2.0 genes in this pathway |
| PID PI3K PLC TRK PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| KIM RESPONSE TO TSA AND DECITABINE UP | 129 | 73 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER GRADES 1 2 UP | 137 | 84 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW UP | 162 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH UP | 147 | 101 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA DN | 181 | 107 | All SZGR 2.0 genes in this pathway |
| MUELLER METHYLATED IN GLIOBLASTOMA | 40 | 22 | All SZGR 2.0 genes in this pathway |
| AIGNER ZEB1 TARGETS | 35 | 15 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| SCHAVOLT TARGETS OF TP53 AND TP63 | 16 | 12 | All SZGR 2.0 genes in this pathway |
| DAUER STAT3 TARGETS UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCIAN CELL CYCLE TARGETS OF TP53 AND TP73 UP | 9 | 7 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS MODERATELY DN | 110 | 64 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| WU CELL MIGRATION | 184 | 114 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA | 164 | 118 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK UP | 87 | 58 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| CROMER METASTASIS DN | 81 | 58 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| MCDOWELL ACUTE LUNG INJURY UP | 45 | 29 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 2 | 50 | 34 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 DN | 65 | 45 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| INGA TP53 TARGETS | 17 | 13 | All SZGR 2.0 genes in this pathway |
| APRELIKOVA BRCA1 TARGETS | 49 | 33 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G1 | 67 | 41 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| HOQUE METHYLATED IN CANCER | 56 | 45 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| HUPER BREAST BASAL VS LUMINAL UP | 54 | 29 | All SZGR 2.0 genes in this pathway |
| HOFMANN MYELODYSPLASTIC SYNDROM LOW RISK DN | 30 | 20 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G23 UP | 52 | 35 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| TERAO AOX4 TARGETS SKIN UP | 38 | 27 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
| KOHOUTEK CCNT1 TARGETS | 50 | 26 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP | 199 | 143 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |