Gene Page: HIF1A
Summary ?
| GeneID | 3091 |
| Symbol | HIF1A |
| Synonyms | HIF-1-alpha|HIF-1A|HIF-1alpha|HIF1|HIF1-ALPHA|MOP1|PASD8|bHLHe78 |
| Description | hypoxia inducible factor 1 alpha subunit |
| Reference | MIM:603348|HGNC:HGNC:4910|Ensembl:ENSG00000100644|HPRD:04517|Vega:OTTHUMG00000140344 |
| Gene type | protein-coding |
| Map location | 14q23.2 |
| Pascal p-value | 0.29 |
| Sherlock p-value | 0.907 |
| TADA p-value | 0.013 |
| Fetal beta | 1.164 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Gulsuner_2013 | Whole Exome Sequencing analysis | 155 DNMs identified by exome sequencing of quads or trios of schizophrenia individuals and their parents. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HIF1A | chr14 | 62203734 | C | G | NM_001243084 NM_001530 NM_181054 | p.410L>V p.386L>V p.386L>V | missense missense missense | Schizophrenia | DNM:Gulsuner_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17139868 | chr7 | 117079228 | HIF1A | 3091 | 0.13 | trans |
Section II. Transcriptome annotation
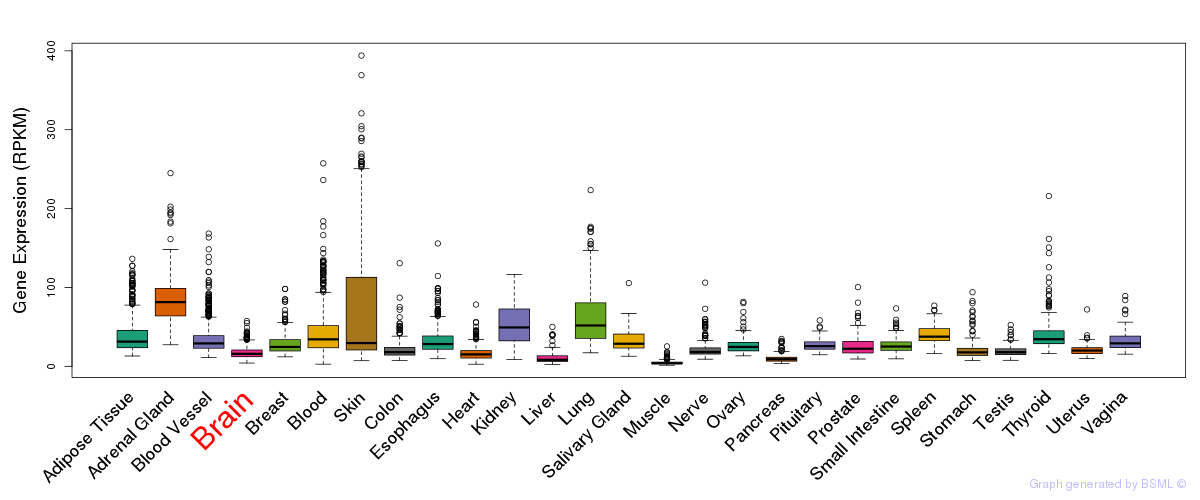
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-DRA | 0.86 | 0.84 |
| HLA-DMA | 0.81 | 0.84 |
| HLA-DPA1 | 0.80 | 0.81 |
| CD74 | 0.78 | 0.81 |
| TBXAS1 | 0.77 | 0.77 |
| CTSS | 0.74 | 0.69 |
| HLA-DRB1 | 0.74 | 0.77 |
| CD86 | 0.73 | 0.64 |
| RGS10 | 0.73 | 0.71 |
| HLA-DQA1 | 0.73 | 0.71 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SH3BP2 | -0.46 | -0.62 |
| KIAA1949 | -0.44 | -0.52 |
| SH2B2 | -0.44 | -0.63 |
| TUBB2B | -0.42 | -0.58 |
| NASP | -0.42 | -0.52 |
| VAV2 | -0.42 | -0.48 |
| ZNF311 | -0.41 | -0.45 |
| ZBTB12 | -0.41 | -0.46 |
| AL033532.1 | -0.41 | -0.48 |
| ZNF300 | -0.41 | -0.44 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APEX1 | APE | APE-1 | APE1 | APEN | APEX | APX | HAP1 | REF-1 | REF1 | APEX nuclease (multifunctional DNA repair enzyme) 1 | - | HPRD,BioGRID | 10594042 |
| ARNT | HIF-1beta | HIF1B | HIF1BETA | TANGO | bHLHe2 | aryl hydrocarbon receptor nuclear translocator | - | HPRD,BioGRID | 9079689 |
| ARNT | HIF-1beta | HIF1B | HIF1BETA | TANGO | bHLHe2 | aryl hydrocarbon receptor nuclear translocator | HIF-1alpha interacts with HIF-1beta | BIND | 8663540 |
| ARNT | HIF-1beta | HIF1B | HIF1BETA | TANGO | bHLHe2 | aryl hydrocarbon receptor nuclear translocator | HIF-1alpha isoform 1 interacts with HIF-1beta isoform 3 | BIND | 8663540 |
| ARNT | HIF-1beta | HIF1B | HIF1BETA | TANGO | bHLHe2 | aryl hydrocarbon receptor nuclear translocator | Arnt interacts with HIF1-alpha. | BIND | 9704006 |
| ARNT | HIF-1beta | HIF1B | HIF1BETA | TANGO | bHLHe2 | aryl hydrocarbon receptor nuclear translocator | HIF-1alpha isoform 2 interacts with HIF-1beta isoform 1 | BIND | 8663540 |
| ARNT | HIF-1beta | HIF1B | HIF1BETA | TANGO | bHLHe2 | aryl hydrocarbon receptor nuclear translocator | HIF-1alpha isoform 2 interacts with HIF-1beta isoform 3 | BIND | 8663540 |
| ARNTL | BMAL1 | BMAL1c | JAP3 | MGC47515 | MOP3 | PASD3 | TIC | bHLHe5 | aryl hydrocarbon receptor nuclear translocator-like | - | HPRD,BioGRID | 9576906 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | HIF-1alpha interacts with CDKN1A promoter. | BIND | 15780936 |
| CDKN2A | ARF | CDK4I | CDKN2 | CMM2 | INK4 | INK4a | MLM | MTS1 | TP16 | p14 | p14ARF | p16 | p16INK4 | p16INK4a | p19 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | p14ARF interacts with HIF-1alpha. | BIND | 11382768 |
| COPS5 | CSN5 | JAB1 | MGC3149 | MOV-34 | SGN5 | COP9 constitutive photomorphogenic homolog subunit 5 (Arabidopsis) | Jab1 interacts with HIF-1alpha. | BIND | 11707426 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD,BioGRID | 10202154 |
| DAP3 | DAP-3 | DKFZp686G12159 | MGC126058 | MGC126059 | MRP-S29 | MRPS29 | bMRP-10 | death associated protein 3 | - | HPRD,BioGRID | 10903152 |
| EGLN1 | C1orf12 | DKFZp761F179 | ECYT3 | HIFPH2 | HPH2 | PHD2 | SM-20 | SM20 | ZMYND6 | egl nine homolog 1 (C. elegans) | HIF-1 alpha interacts with PHD2. | BIND | 15721254 |
| EGLN3 | FLJ21620 | HIFPH3 | MGC125998 | MGC125999 | PHD3 | egl nine homolog 3 (C. elegans) | HIF-1 alpha interacts with PHD3. | BIND | 15721254 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | - | HPRD,BioGRID | 11823643 |
| EPO | EP | MGC138142 | erythropoietin | HIF1A (HIF-1-alpha) interacts with the EPO gene 3(prime) enhancer. | BIND | 15674338 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | Reconstituted Complex | BioGRID | 11641274 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Reconstituted Complex | BioGRID | 11641274 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | Reconstituted Complex | BioGRID | 11641274 |
| HIF1AN | DKFZp762F1811 | FIH1 | FLJ20615 | FLJ22027 | hypoxia-inducible factor 1, alpha subunit inhibitor | - | HPRD,BioGRID | 11641274 |
| HIF3A | HIF-3A | HIF-3A2 | HIF-3A4 | IPAS | MOP7 | PASD7 | bHLHe17 | hypoxia inducible factor 3, alpha subunit | - | HPRD | 11734856 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD,BioGRID | 9079689 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 11739718 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | - | HPRD | 10551817 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 10640274 |12606552 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | - | HPRD | 10640274|12606552 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD,BioGRID | 10594042 |
| OS9 | - | amplified in osteosarcoma | HIF-1 alpha interacts with OS-9. | BIND | 15721254 |
| PER1 | MGC88021 | PER | RIGUI | hPER | period homolog 1 (Drosophila) | - | HPRD,BioGRID | 11726537 |
| PGK1 | MGC117307 | MGC142128 | MGC8947 | MIG10 | PGKA | phosphoglycerate kinase 1 | - | HPRD | 8089148 |
| PSMA7 | C6 | HSPC | MGC3755 | RC6-1 | XAPC7 | proteasome (prosome, macropain) subunit, alpha type, 7 | - | HPRD,BioGRID | 11389899 |
| PSMC3 | MGC8487 | TBP1 | proteasome (prosome, macropain) 26S subunit, ATPase, 3 | - | HPRD,BioGRID | 14556007 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | RB1 (pRB) interacts with HIF1A (HIF-1-alpha). | BIND | 15674338 |
| SP1 | - | Sp1 transcription factor | Sp1 interacts with HIF-1alpha. | BIND | 15780936 |
| SSX4 | MGC119056 | MGC12411 | synovial sarcoma, X breakpoint 4 | Two-hybrid | BioGRID | 16189514 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9537326 |10640274 |12124396 |12606552 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD | 9537326|12124396 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with HIF-1 alpha. | BIND | 15629713 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | HIF-1alpha interacts with p53 | BIND | 11593383 |
| VEGFA | MGC70609 | VEGF | VEGF-A | VPF | vascular endothelial growth factor A | HIF1A (HIF-1-alpha) interacts with the VEGF promoter. | BIND | 15674338 |
| VHL | HRCA1 | RCA1 | VHL1 | von Hippel-Lindau tumor suppressor | Hydroxylated HIF-1 alpha interacts with VHL. This interaction was modeled on a demonstrated interaction between human hydroxylated HIF-1 alpha and an unspecified isoform of VHL from an unspecified species. | BIND | 15721254 |
| VHL | HRCA1 | RCA1 | VHL1 | von Hippel-Lindau tumor suppressor | The von Hippel-Lindau tumor suppressor protein (pVHL) interacts with Hypoxia inducible factor-alpha (HIF-1 alpha) | BIND | 10823831 |
| VHL | HRCA1 | RCA1 | VHL1 | von Hippel-Lindau tumor suppressor | - | HPRD | 11504942 |
| VHL | HRCA1 | RCA1 | VHL1 | von Hippel-Lindau tumor suppressor | HIF1A (HIF-1alpha) interacts with VHL. | BIND | 15750626 |
| VHL | HRCA1 | RCA1 | VHL1 | von Hippel-Lindau tumor suppressor | - | HPRD,BioGRID | 10944113 |
| VHLL | VLP | von Hippel-Lindau tumor suppressor-like | - | HPRD | 14757845 |
| ZNF197 | D3S1363E | P18 | VHLaK | ZKSCAN9 | ZNF166 | ZNF20 | zinc finger protein 197 | Affinity Capture-Western | BioGRID | 12682018 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MTOR SIGNALING PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG RENAL CELL CARCINOMA | 70 | 60 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EPONFKB PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P53HYPOXIA PATHWAY | 23 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIF PATHWAY | 15 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VEGF PATHWAY | 29 | 18 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID HIF1A PATHWAY | 19 | 12 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| PID HIF1 TFPATHWAY | 66 | 52 | All SZGR 2.0 genes in this pathway |
| PID HES HEY PATHWAY | 48 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | 46 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH1 | 70 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | 18 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME CIRCADIAN CLOCK | 53 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN | 186 | 107 | All SZGR 2.0 genes in this pathway |
| MARKS HDAC TARGETS DN | 15 | 11 | All SZGR 2.0 genes in this pathway |
| BAKER HEMATOPOIESIS STAT3 TARGETS | 16 | 12 | All SZGR 2.0 genes in this pathway |
| MARTIN INTERACT WITH HDAC | 44 | 31 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 4NM DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| HASINA NOL7 TARGETS UP | 13 | 11 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS MODERATELY DN | 110 | 64 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS SUBSET | 33 | 20 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| ROSS ACUTE MYELOID LEUKEMIA CBF | 82 | 57 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION UP | 71 | 51 | All SZGR 2.0 genes in this pathway |
| DER IFN BETA RESPONSE UP | 102 | 67 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| HARRIS HYPOXIA | 81 | 64 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C2 | 54 | 39 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS UP | 77 | 57 | All SZGR 2.0 genes in this pathway |
| KEEN RESPONSE TO ROSIGLITAZONE DN | 106 | 68 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| HOWLIN CITED1 TARGETS 1 DN | 37 | 22 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN THYMUS UP | 196 | 137 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL DN | 175 | 103 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| ROSS LEUKEMIA WITH MLL FUSIONS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS ONCOGENIC SIGNATURE | 89 | 56 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| VALK AML WITH 11Q23 REARRANGED | 22 | 13 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA LB DN | 44 | 23 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 11 | 103 | 68 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 DN | 157 | 106 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| RAFFEL VEGFA TARGETS UP | 9 | 8 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR133 TARGETS UP | 43 | 27 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |