Gene Page: KPNB1
Summary ?
| GeneID | 3837 |
| Symbol | KPNB1 |
| Synonyms | IMB1|IPO1|IPOB|Impnb|NTF97 |
| Description | karyopherin subunit beta 1 |
| Reference | MIM:602738|HGNC:HGNC:6400|HPRD:04114| |
| Gene type | protein-coding |
| Map location | 17q21.32 |
| Pascal p-value | 0.595 |
| Sherlock p-value | 0.513 |
| Fetal beta | 0.112 |
| DMG | 2 (# studies) |
| Support | STRUCTURAL PLASTICITY CompositeSet Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| Expression | Meta-analysis of gene expression | P value: 2.337 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02415717 | 17 | 45727607 | KPNB1 | 3.865E-4 | -0.253 | 0.043 | DMG:Wockner_2014 |
| cg24438655 | 17 | 45727598 | KPNB1 | -0.019 | 0.61 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
General gene expression (GTEx)
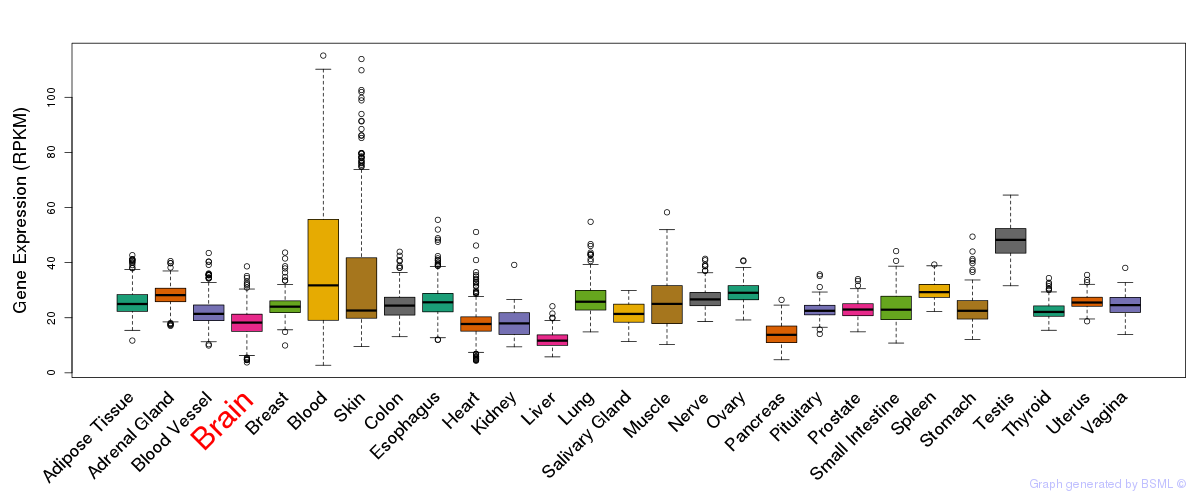
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008270 | zinc ion binding | TAS | 7615630 | |
| GO:0008565 | protein transporter activity | IEA | - | |
| GO:0008139 | nuclear localization sequence binding | TAS | 7627554 | |
| GO:0019904 | protein domain specific binding | IPI | 11984006 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000059 | protein import into nucleus, docking | IEA | - | |
| GO:0000060 | protein import into nucleus, translocation | TAS | 7615630 | |
| GO:0006607 | NLS-bearing substrate import into nucleus | TAS | 7627554 | |
| GO:0006610 | ribosomal protein import into nucleus | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 1985200 |7559393 |9405152 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005635 | nuclear envelope | IEA | - | |
| GO:0005643 | nuclear pore | TAS | 7627554 | |
| GO:0005654 | nucleoplasm | EXP | 1985200 |7559393 |9405152 | |
| GO:0005737 | cytoplasm | TAS | 7615630 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CD99 | MIC2 | MIC2X | MIC2Y | CD99 molecule | CD99 interacts with KPNB1. | BIND | 15388255 |
| ELAVL1 | ELAV1 | HUR | Hua | MelG | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 1 (Hu antigen R) | - | HPRD | 15037768 |15342649 |
| FGFR1 | BFGFR | CD331 | CEK | FGFBR | FLG | FLJ99988 | FLT2 | HBGFR | KAL2 | N-SAM | fibroblast growth factor receptor 1 | - | HPRD,BioGRID | 11257130 |
| H1F0 | H10 | H1FV | MGC5241 | H1 histone family, member 0 | in vitro | BioGRID | 10228156 |
| IPO7 | FLJ14581 | Imp7 | MGC138673 | RANBP7 | importin 7 | - | HPRD,BioGRID | 9214382 |10228156 |
| IPO8 | FLJ26580 | RANBP8 | importin 8 | - | HPRD,BioGRID | 9214382 |
| KPNA1 | IPOA5 | NPI-1 | RCH2 | SRP1 | karyopherin alpha 1 (importin alpha 5) | Reconstituted Complex | BioGRID | 9102465 |10523667 |
| KPNA2 | IPOA1 | QIP2 | RCH1 | SRP1alpha | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | Reconstituted Complex | BioGRID | 9323134 |10523667 |11283257 |
| KPNA2 | IPOA1 | QIP2 | RCH1 | SRP1alpha | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | - | HPRD | 8617226 |
| KPNA3 | IPOA4 | SRP1gamma | SRP4 | hSRP1 | karyopherin alpha 3 (importin alpha 4) | Reconstituted Complex | BioGRID | 9435235 |10523667 |
| KPNA4 | IPOA3 | MGC12217 | MGC26703 | QIP1 | SRP3 | karyopherin alpha 4 (importin alpha 3) | Reconstituted Complex | BioGRID | 10523667 |
| KPNA6 | FLJ11249 | IPOA7 | KPNA7 | MGC17918 | karyopherin alpha 6 (importin alpha 7) | - | HPRD,BioGRID | 10523667 |
| LYST | CHS | CHS1 | lysosomal trafficking regulator | - | HPRD,BioGRID | 11984006 |
| NUP153 | HNUP153 | N153 | nucleoporin 153kDa | - | HPRD | 9275187|10749866 |
| NUP153 | HNUP153 | N153 | nucleoporin 153kDa | Reconstituted Complex | BioGRID | 10202161 |10749866 |11266456 |
| NUP50 | MGC39961 | NPAP60 | NPAP60L | nucleoporin 50kDa | - | HPRD,BioGRID | 12176322 |
| NUP54 | MGC13407 | nucleoporin 54kDa | Reconstituted Complex | BioGRID | 11266456 |
| NUP62 | DKFZp547L134 | FLJ20822 | FLJ43869 | IBSN | MGC841 | SNDI | p62 | nucleoporin 62kDa | Reconstituted Complex | BioGRID | 9102465 |11266456 |
| NUP98 | ADIR2 | NUP196 | NUP96 | nucleoporin 98kDa | - | HPRD,BioGRID | 9144189 |
| NUTF2 | NTF2 | PP15 | nuclear transport factor 2 | Reconstituted Complex | BioGRID | 9102465 |
| PTHLH | HHM | MGC14611 | PLP | PTHR | PTHRP | parathyroid hormone-like hormone | - | HPRD,BioGRID | 11401507 |12504010 |
| PTMA | MGC104802 | TMSA | prothymosin, alpha | - | HPRD | 11310559 |
| RAN | ARA24 | Gsp1 | TC4 | RAN, member RAS oncogene family | Far Western Reconstituted Complex | BioGRID | 9102465 |9135132 |10779340 |
| RAN | ARA24 | Gsp1 | TC4 | RAN, member RAS oncogene family | - | HPRD | 8576188 |9135132 |
| RAN | ARA24 | Gsp1 | TC4 | RAN, member RAS oncogene family | Interaction between KPNB1 (PDB ID: 1IBR_B) and RAN (PDB ID: 1IBR_A). | BIND | 7615630 |7627554 |8896452 |10367892 |
| RANBP1 | HTF9A | MGC88701 | RAN binding protein 1 | Reconstituted Complex | BioGRID | 10473610 |10779340 |
| RANBP2 | NUP358 | TRP1 | TRP2 | RAN binding protein 2 | - | HPRD,BioGRID | 10473610 |
| RUNX1T1 | AML1T1 | CBFA2T1 | CDR | ETO | MGC2796 | MTG8 | MTG8b | ZMYND2 | runt-related transcription factor 1; translocated to, 1 (cyclin D-related) | - | HPRD,BioGRID | 10951564 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD,BioGRID | 10846168 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | Affinity Capture-Western | BioGRID | 10846168 |
| SMN1 | BCD541 | SMA | SMA1 | SMA2 | SMA3 | SMA4 | SMA@ | SMN | SMNT | T-BCD541 | survival of motor neuron 1, telomeric | - | HPRD,BioGRID | 12095920 |
| SNUPN | KPNBL | RNUT1 | Snurportin1 | snurportin 1 | - | HPRD | 12095920 |
| SOX9 | CMD1 | CMPD1 | SRA1 | SRY (sex determining region Y)-box 9 | KPNB1 (importin beta) interacts with SOX9. | BIND | 15889150 |
| SOX9 | CMD1 | CMPD1 | SRA1 | SRY (sex determining region Y)-box 9 | - | HPRD | 11323423 |
| SREBF2 | SREBP2 | bHLHd2 | sterol regulatory element binding transcription factor 2 | Reconstituted Complex | BioGRID | 11283257 |
| TERF1 | FLJ41416 | PIN2 | TRBF1 | TRF | TRF1 | hTRF1-AS | t-TRF1 | telomeric repeat binding factor (NIMA-interacting) 1 | Reconstituted Complex | BioGRID | 12135354 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 11297531 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD | 14743216 |
| UBR5 | DD5 | EDD | EDD1 | FLJ11310 | HYD | KIAA0896 | MGC57263 | ubiquitin protein ligase E3 component n-recognin 5 | - | HPRD | 12011095 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA RANMS PATHWAY | 10 | 7 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID TCPTP PATHWAY | 43 | 33 | All SZGR 2.0 genes in this pathway |
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3PATHWAY | 17 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | 66 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | 13 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA LIFE CYCLE | 203 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | 27 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME HOST INTERACTIONS OF HIV FACTORS | 132 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC EXECUTION PHASE | 54 | 37 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA DN | 70 | 38 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| TIMOFEEVA GROWTH STRESS VIA STAT1 DN | 16 | 12 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| HWANG PROSTATE CANCER MARKERS | 28 | 19 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA PROGRESSION RISK | 74 | 44 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER UP | 181 | 108 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR ANAPLASTIC UP | 19 | 12 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| LIU BREAST CANCER | 30 | 19 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION DN | 87 | 57 | All SZGR 2.0 genes in this pathway |
| WANG TARGETS OF MLL CBP FUSION DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| PARK HSC AND MULTIPOTENT PROGENITORS | 50 | 33 | All SZGR 2.0 genes in this pathway |
| GALE APL WITH FLT3 MUTATED UP | 56 | 35 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION MUSCLE DN | 51 | 28 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA E2 | 118 | 65 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| KAPOSI LIVER CANCER MET UP | 18 | 11 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 2 | 86 | 50 | All SZGR 2.0 genes in this pathway |
| KAMMINGA SENESCENCE | 41 | 26 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| WIERENGA PML INTERACTOME | 42 | 23 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |
| ABRAMSON INTERACT WITH AIRE | 45 | 33 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 492 | 499 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-145 | 36 | 42 | 1A | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-148/152 | 207 | 213 | 1A | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-181 | 876 | 883 | 1A,m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-182 | 1093 | 1099 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-194 | 953 | 959 | 1A | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-199 | 35 | 41 | 1A | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-203.1 | 635 | 641 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-218 | 673 | 679 | 1A | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-27 | 872 | 879 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-342 | 554 | 560 | m8 | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
| miR-361 | 1208 | 1215 | 1A,m8 | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
| miR-377 | 553 | 559 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-9 | 1095 | 1101 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-96 | 1092 | 1099 | 1A,m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.