| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP
| 285 | 181 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN
| 481 | 290 | All SZGR 2.0 genes in this pathway |
| KIM RESPONSE TO TSA AND DECITABINE UP
| 129 | 73 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN
| 349 | 157 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN
| 391 | 222 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 DN
| 175 | 82 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN
| 198 | 110 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP
| 430 | 232 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN
| 260 | 143 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP
| 171 | 112 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP
| 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN
| 805 | 505 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP
| 530 | 342 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP
| 215 | 137 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP
| 214 | 144 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP
| 271 | 175 | All SZGR 2.0 genes in this pathway |
| OUELLET CULTURED OVARIAN CANCER INVASIVE VS LMP UP
| 69 | 40 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER
| 178 | 108 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL
| 326 | 213 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL
| 330 | 217 | All SZGR 2.0 genes in this pathway |
| TSUNODA CISPLATIN RESISTANCE DN
| 51 | 38 | All SZGR 2.0 genes in this pathway |
| KIM MYCN AMPLIFICATION TARGETS DN
| 103 | 59 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS
| 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP
| 487 | 286 | All SZGR 2.0 genes in this pathway |
| WU CELL MIGRATION
| 184 | 114 | All SZGR 2.0 genes in this pathway |
| LIN SILENCED BY TUMOR MICROENVIRONMENT
| 108 | 73 | All SZGR 2.0 genes in this pathway |
| KENNY CTNNB1 TARGETS UP
| 50 | 30 | All SZGR 2.0 genes in this pathway |
| ZUCCHI METASTASIS DN
| 44 | 29 | All SZGR 2.0 genes in this pathway |
| SANA TNF SIGNALING DN
| 90 | 57 | All SZGR 2.0 genes in this pathway |
| CHEN LUNG CANCER SURVIVAL
| 28 | 22 | All SZGR 2.0 genes in this pathway |
| LIN APC TARGETS
| 77 | 55 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO ESTRADIOL DN
| 82 | 52 | All SZGR 2.0 genes in this pathway |
| SANA RESPONSE TO IFNG DN
| 85 | 56 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP
| 157 | 105 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR DN
| 25 | 15 | All SZGR 2.0 genes in this pathway |
| GILDEA METASTASIS
| 30 | 14 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS
| 318 | 215 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD DN
| 162 | 102 | All SZGR 2.0 genes in this pathway |
| PETROVA PROX1 TARGETS DN
| 64 | 38 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP
| 101 | 76 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM
| 302 | 191 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN
| 830 | 547 | All SZGR 2.0 genes in this pathway |
| VIETOR IFRD1 TARGETS
| 23 | 13 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA UP
| 52 | 33 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS
| 108 | 71 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 UP
| 84 | 55 | All SZGR 2.0 genes in this pathway |
| LIANG SILENCED BY METHYLATION UP
| 32 | 18 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1
| 419 | 273 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 3HR
| 74 | 47 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP
| 282 | 183 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP
| 578 | 341 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN
| 270 | 181 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN
| 564 | 326 | All SZGR 2.0 genes in this pathway |
| MISHRA CARCINOMA ASSOCIATED FIBROBLAST DN
| 24 | 13 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP
| 425 | 298 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS DN
| 39 | 20 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN
| 240 | 153 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP
| 491 | 316 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE UP
| 91 | 63 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS FIBROBLAST UP
| 84 | 60 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED
| 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3
| 269 | 159 | All SZGR 2.0 genes in this pathway |
| LIU VAV3 PROSTATE CARCINOGENESIS UP
| 89 | 61 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL UP
| 73 | 49 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER HEPATOBLAST
| 16 | 12 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3
| 435 | 318 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 1
| 68 | 50 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP
| 387 | 225 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 1 UP
| 7 | 5 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP
| 745 | 475 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3
| 824 | 528 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS UP
| 221 | 120 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN
| 211 | 119 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP
| 289 | 184 | All SZGR 2.0 genes in this pathway |
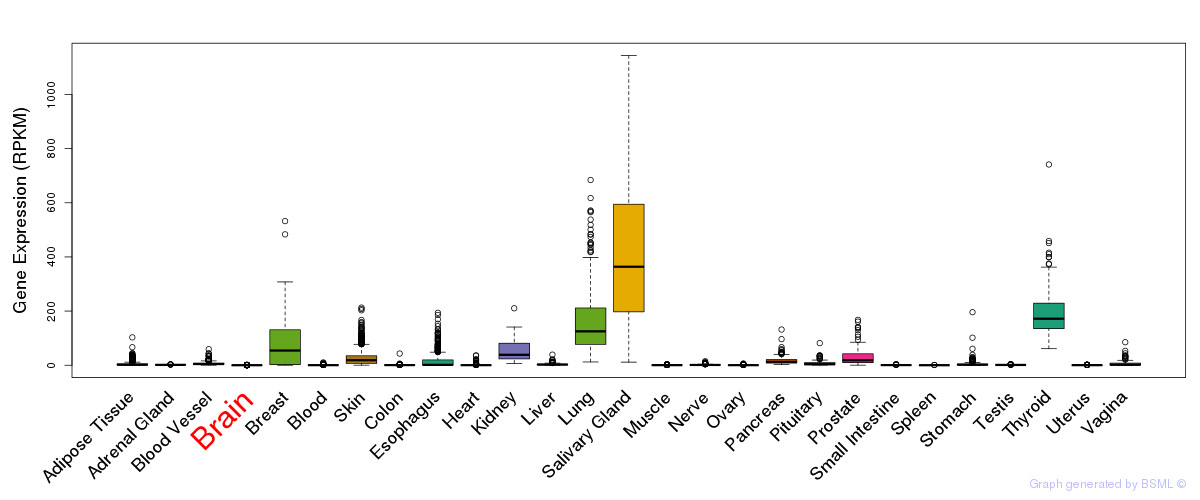
 Differentially methylated gene
Differentially methylated gene