Gene Page: KTN1
Summary ?
| GeneID | 3895 |
| Symbol | KTN1 |
| Synonyms | CG1|KNT|MU-RMS-40.19 |
| Description | kinectin 1 |
| Reference | MIM:600381|HGNC:HGNC:6467|Ensembl:ENSG00000126777|HPRD:02661|Vega:OTTHUMG00000140312 |
| Gene type | protein-coding |
| Map location | 14q22.1 |
| Pascal p-value | 0.07 |
| Sherlock p-value | 0.967 |
| Fetal beta | -0.692 |
| eGene | Myers' cis & trans |
| Support | INTRACELLULAR TRAFFICKING G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs946067 | chr14 | 55932549 | KTN1 | 3895 | 0.11 | cis | ||
| rs11158041 | chr14 | 55934219 | KTN1 | 3895 | 0.01 | cis | ||
| rs11158043 | chr14 | 55934726 | KTN1 | 3895 | 0.03 | cis | ||
| rs3742571 | chr14 | 56016703 | KTN1 | 3895 | 0.01 | cis | ||
| rs10142497 | chr14 | 56047947 | KTN1 | 3895 | 1.738E-5 | cis | ||
| rs10137340 | chr14 | 56050988 | KTN1 | 3895 | 7.702E-6 | cis | ||
| rs17683656 | chr14 | 56053360 | KTN1 | 3895 | 3.871E-4 | cis | ||
| rs8017891 | chr14 | 56172966 | KTN1 | 3895 | 4.402E-5 | cis | ||
| rs17832389 | chr14 | 56185835 | KTN1 | 3895 | 1.005E-4 | cis | ||
| rs10142497 | chr14 | 56047947 | KTN1 | 3895 | 0 | trans | ||
| rs10137340 | chr14 | 56050988 | KTN1 | 3895 | 0 | trans | ||
| rs17683656 | chr14 | 56053360 | KTN1 | 3895 | 0.03 | trans | ||
| rs8017891 | chr14 | 56172966 | KTN1 | 3895 | 0.01 | trans | ||
| rs17832389 | chr14 | 56185835 | KTN1 | 3895 | 0.01 | trans |
Section II. Transcriptome annotation
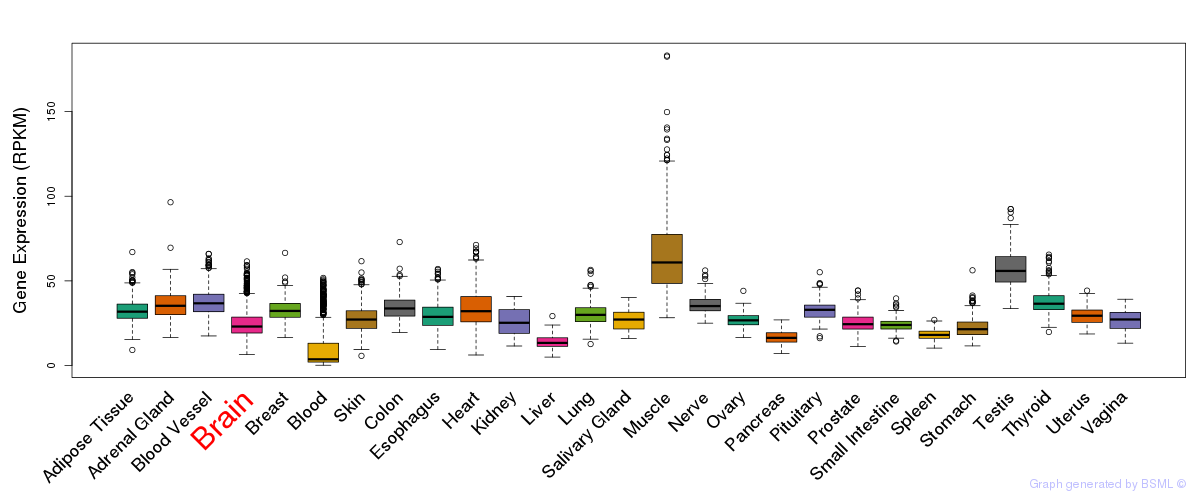
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RNF13 | 0.95 | 0.93 |
| ABCA8 | 0.94 | 0.86 |
| ANLN | 0.94 | 0.77 |
| LIPA | 0.94 | 0.94 |
| PRRG1 | 0.94 | 0.87 |
| EDIL3 | 0.93 | 0.87 |
| CDH19 | 0.93 | 0.81 |
| UGT8 | 0.93 | 0.87 |
| PLD1 | 0.93 | 0.84 |
| ENDOD1 | 0.93 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TBC1D10A | -0.52 | -0.47 |
| BCL7C | -0.50 | -0.66 |
| SH3BP2 | -0.49 | -0.63 |
| SH2B2 | -0.49 | -0.64 |
| CCDC28B | -0.48 | -0.64 |
| NR2C2AP | -0.48 | -0.52 |
| PLEKHO1 | -0.48 | -0.53 |
| MMP25 | -0.47 | -0.52 |
| KIAA1949 | -0.47 | -0.46 |
| TRAF4 | -0.47 | -0.58 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM UP | 98 | 58 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 DN | 116 | 71 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK UP | 116 | 68 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| XU CREBBP TARGETS UP | 26 | 16 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL CARCINOMA VS ADENOMA DN | 24 | 18 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN ACTIVATED B LYMPHOCYTE | 81 | 66 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING UP | 93 | 62 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |