Gene Page: LRCH4
Summary ?
| GeneID | 4034 |
| Symbol | LRCH4 |
| Synonyms | LRN|LRRN1|LRRN4|PP14183 |
| Description | leucine-rich repeats and calponin homology (CH) domain containing 4 |
| Reference | HGNC:HGNC:6691|HPRD:10062| |
| Gene type | protein-coding |
| Map location | 7q22 |
| Pascal p-value | 0.042 |
| DEG p-value | DEG:Maycox_2009:CC_BA10_fold_change=1.20:CC_BA10_disease_P=0.0151:HBB_BA9_fold_change=1.15:HBB_BA9_disease_P=0.0117 |
| eGene | Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Maycox_2009 | Microarray to determine the expression of over 30000 mRNA transcripts in post-mortem tissue | We included 51 genes whose expression changes are common between two schizophrenia cohorts. | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | LRCH4 | 4034 | 0 | trans | ||
| rs7584986 | chr2 | 184111432 | LRCH4 | 4034 | 0.19 | trans | ||
| rs16890367 | chr6 | 38078448 | LRCH4 | 4034 | 0.01 | trans | ||
| rs16955618 | chr15 | 29937543 | LRCH4 | 4034 | 3.245E-6 | trans |
Section II. Transcriptome annotation
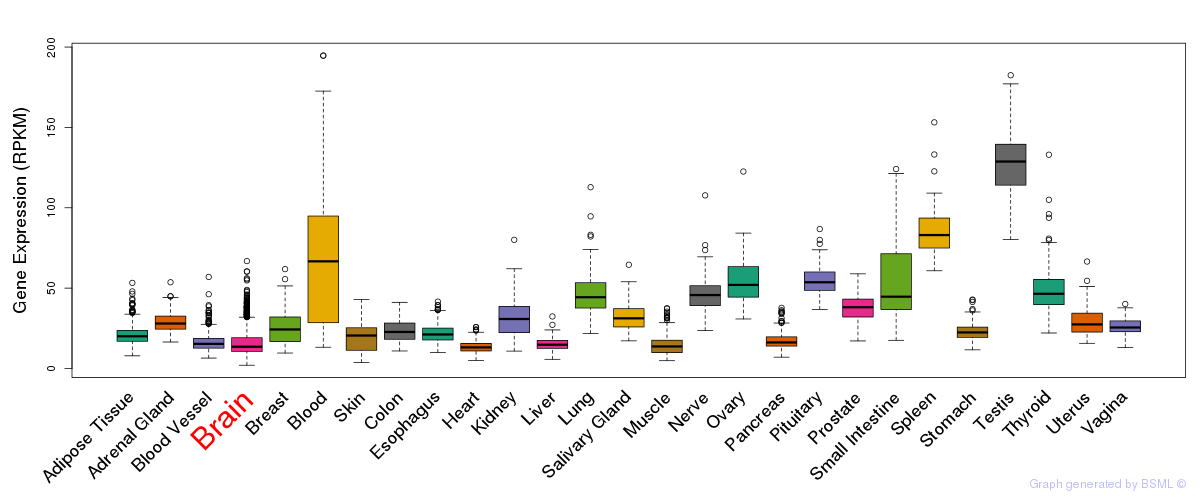
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-E | 0.82 | 0.85 |
| GGT5 | 0.81 | 0.83 |
| TINAGL1 | 0.79 | 0.79 |
| TBX2 | 0.78 | 0.81 |
| GPER | 0.78 | 0.82 |
| LSR | 0.77 | 0.79 |
| FGR | 0.77 | 0.78 |
| C1QTNF1 | 0.77 | 0.80 |
| A4GALT | 0.76 | 0.77 |
| GRIN2C | 0.76 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FRG1 | -0.66 | -0.73 |
| COQ3 | -0.63 | -0.65 |
| C18orf21 | -0.62 | -0.65 |
| GNL3 | -0.61 | -0.63 |
| DNAJC2 | -0.61 | -0.66 |
| C11orf57 | -0.61 | -0.71 |
| PAK1IP1 | -0.60 | -0.63 |
| HELQ | -0.60 | -0.59 |
| SUPV3L1 | -0.60 | -0.64 |
| SRP19 | -0.60 | -0.58 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| GO:0016564 | transcription repressor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 9799793 |
| GO:0016481 | negative regulation of transcription | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016605 | PML body | IDA | 16449650 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| SCIBETTA KDM5B TARGETS DN | 81 | 55 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN | 245 | 144 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN WITHOUT MGMT 48HR DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP | 253 | 147 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 8G | 95 | 62 | All SZGR 2.0 genes in this pathway |
| ENGELMANN CANCER PROGENITORS DN | 70 | 44 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| SENGUPTA EBNA1 ANTICORRELATED | 173 | 85 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 80 | 86 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-503 | 819 | 825 | m8 | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
| miR-9 | 95 | 102 | 1A,m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.