Gene Page: ARNT
Summary ?
| GeneID | 405 |
| Symbol | ARNT |
| Synonyms | HIF-1-beta|HIF-1beta|HIF1-beta|HIF1B|HIF1BETA|TANGO|bHLHe2 |
| Description | aryl hydrocarbon receptor nuclear translocator |
| Reference | MIM:126110|HGNC:HGNC:700|Ensembl:ENSG00000143437|HPRD:00524|Vega:OTTHUMG00000035011 |
| Gene type | protein-coding |
| Map location | 1q21 |
| Sherlock p-value | 0.87 |
| Fetal beta | -0.586 |
| eGene | Caudate basal ganglia Cerebellum Cortex |
| Support | G2Cdb.humanNRC G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
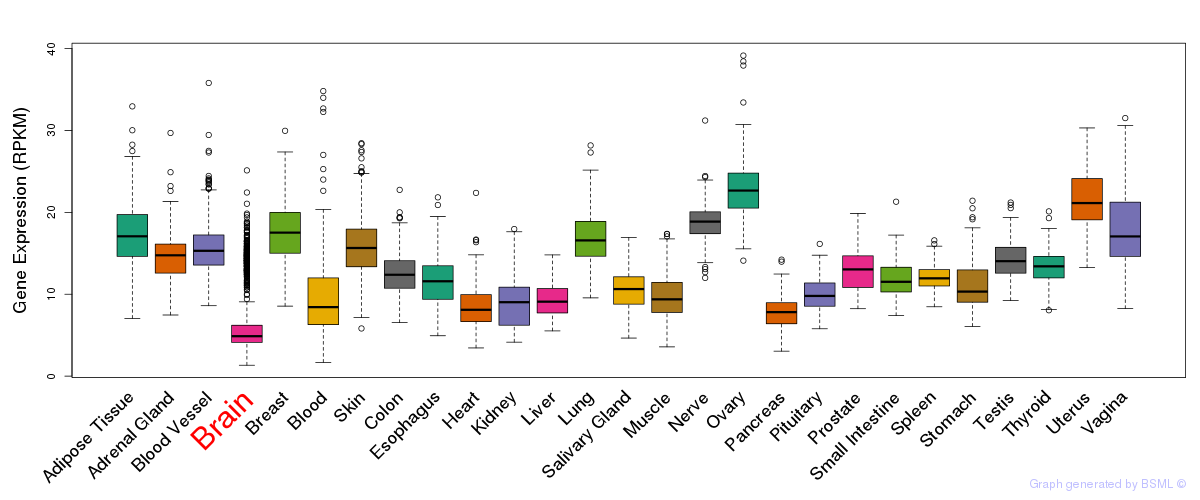
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AL117209.1 | 0.86 | 0.85 |
| MAGI2 | 0.85 | 0.85 |
| TATDN2 | 0.85 | 0.83 |
| INPP4A | 0.84 | 0.86 |
| BRPF3 | 0.84 | 0.85 |
| MAPRE2 | 0.84 | 0.84 |
| ATCAY | 0.84 | 0.86 |
| MSMP | 0.84 | 0.85 |
| DNAJC5 | 0.84 | 0.86 |
| RGP1 | 0.84 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.68 | -0.62 |
| GNG11 | -0.65 | -0.63 |
| HIGD1B | -0.64 | -0.61 |
| AF347015.31 | -0.63 | -0.59 |
| MT-CO2 | -0.63 | -0.58 |
| C1orf54 | -0.62 | -0.67 |
| AL138743.2 | -0.62 | -0.64 |
| AP002478.3 | -0.60 | -0.59 |
| IL32 | -0.59 | -0.53 |
| NOSTRIN | -0.59 | -0.54 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004871 | signal transducer activity | IEA | - | |
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0003705 | RNA polymerase II transcription factor activity, enhancer binding | IDA | 7539918 |8756616 | |
| GO:0003713 | transcription coactivator activity | IEA | - | |
| GO:0003713 | transcription coactivator activity | TAS | 1317062 | |
| GO:0017162 | aryl hydrocarbon receptor binding | IPI | 9079689 | |
| GO:0046982 | protein heterodimerization activity | IEA | - | |
| GO:0046982 | protein heterodimerization activity | IPI | 9079689 | |
| GO:0043565 | sequence-specific DNA binding | IDA | 7539918 |8756616 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001666 | response to hypoxia | IDA | 8756616 | |
| GO:0001892 | embryonic placenta development | IEA | - | |
| GO:0001938 | positive regulation of endothelial cell proliferation | IC | 8756616 | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0010575 | positive regulation vascular endothelial growth factor production | IDA | 8756616 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0043193 | positive regulation of gene-specific transcription | IDA | 8089148 | |
| GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway | IC | 8756616 | |
| GO:0042789 | mRNA transcription from RNA polymerase II promoter | IC | 7539918 | |
| GO:0045648 | positive regulation of erythrocyte differentiation | IC | 1448077 | |
| GO:0045941 | positive regulation of transcription | IEA | - | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0046886 | positive regulation of hormone biosynthetic process | IDA | 1448077 | |
| GO:0045821 | positive regulation of glycolysis | IC | 8089148 | |
| GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress | IDA | 8089148 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005634 | nucleus | TAS | 1317062 | |
| GO:0005667 | transcription factor complex | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AHR | - | aryl hydrocarbon receptor | - | HPRD | 7488247 |7628454 |8384309 |11768231 |12024042 |
| AHR | - | aryl hydrocarbon receptor | Arnt interacts with AhR. | BIND | 9704006 |
| AHR | - | aryl hydrocarbon receptor | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 7488247 |7628454 |8384309 |9887096 |
| AHRR | AHH | AHHR | KIAA1234 | MGC167813 | MGC176630 | aryl-hydrocarbon receptor repressor | - | HPRD,BioGRID | 9887096 |
| AIP | ARA9 | FKBP16 | FKBP37 | SMTPHN | XAP2 | aryl hydrocarbon receptor interacting protein | - | HPRD,BioGRID | 9111057 |
| ARNT | HIF-1beta | HIF1B | HIF1BETA | TANGO | bHLHe2 | aryl hydrocarbon receptor nuclear translocator | Arnt interacts with another copy of itself, albeit weakly, to form a homodimer. | BIND | 9704006 |
| EPAS1 | ECYT4 | HIF2A | HLF | MOP2 | PASD2 | endothelial PAS domain protein 1 | HLF interacts with Arnt. This interaction was modeled on a demonstrated interaction between mouse HLF and human Arnt. | BIND | 9113979 |
| EPAS1 | ECYT4 | HIF2A | HLF | MOP2 | PASD2 | endothelial PAS domain protein 1 | - | HPRD,BioGRID | 9079689 |
| EPAS1 | ECYT4 | HIF2A | HLF | MOP2 | PASD2 | endothelial PAS domain protein 1 | Arnt interacts with HLF. This interaction was modeled on a demonstrated interaction between human Arnt and mouse HLF. | BIND | 9704006 |
| GTF2F1 | BTF4 | RAP74 | TF2F1 | TFIIF | general transcription factor IIF, polypeptide 1, 74kDa | Reconstituted Complex | BioGRID | 8794892 |
| GTF2F2 | BTF4 | RAP30 | TF2F2 | TFIIF | general transcription factor IIF, polypeptide 2, 30kDa | - | HPRD,BioGRID | 8794892 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | - | HPRD,BioGRID | 9079689 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | HIF-1alpha interacts with HIF-1beta | BIND | 8663540 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | HIF-1alpha isoform 1 interacts with HIF-1beta isoform 3 | BIND | 8663540 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | Arnt interacts with HIF1-alpha. | BIND | 9704006 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | HIF-1alpha isoform 2 interacts with HIF-1beta isoform 1 | BIND | 8663540 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | HIF-1alpha isoform 2 interacts with HIF-1beta isoform 3 | BIND | 8663540 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | Affinity Capture-Western | BioGRID | 11013261 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD | 10594042 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | - | HPRD,BioGRID | 12024042 |
| NPAS1 | MOP5 | PASD5 | bHLHe11 | neuronal PAS domain protein 1 | NPAS1 interacts with Arnt | BIND | 15635607 |
| NPAS4 | Le-PAS | NXF | PASD10 | neuronal PAS domain protein 4 | - | HPRD,BioGRID | 14701734 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD,BioGRID | 12354770 |
| PTGES3 | P23 | TEBP | cPGES | prostaglandin E synthase 3 (cytosolic) | - | HPRD | 11879970 |
| PTGES3 | P23 | TEBP | cPGES | prostaglandin E synthase 3 (cytosolic) | Reconstituted Complex | BioGRID | 11259606 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Affinity Capture-Western | BioGRID | 9712901 |
| SIM1 | bHLHe14 | single-minded homolog 1 (Drosophila) | - | HPRD,BioGRID | 9020169 |
| SIM2 | MGC119447 | SIM | bHLHe15 | single-minded homolog 2 (Drosophila) | - | HPRD,BioGRID | 9020169 |9271372 |11782478 |
| SP1 | - | Sp1 transcription factor | Reconstituted Complex | BioGRID | 10471301 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12024042 |
| TAZ | BTHS | CMD3A | EFE | EFE2 | FLJ27390 | G4.5 | LVNCX | Taz1 | XAP-2 | tafazzin | Affinity Capture-Western | BioGRID | 12065584 |
| XPO1 | CRM1 | DKFZp686B1823 | exportin 1 (CRM1 homolog, yeast) | Affinity Capture-Western | BioGRID | 12065584 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG RENAL CELL CARCINOMA | 70 | 60 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EPONFKB PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIF PATHWAY | 15 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VEGF PATHWAY | 29 | 18 | All SZGR 2.0 genes in this pathway |
| PID HIF2PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID HIF1A PATHWAY | 19 | 12 | All SZGR 2.0 genes in this pathway |
| PID HIF1 TFPATHWAY | 66 | 52 | All SZGR 2.0 genes in this pathway |
| PID HES HEY PATHWAY | 48 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | 25 | 17 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED UP | 78 | 48 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION UP | 119 | 66 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP | 215 | 137 | All SZGR 2.0 genes in this pathway |
| STREICHER LSM1 TARGETS UP | 44 | 34 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 17 | 25 | 12 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 24 | 14 | 7 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BYSTROEM CORRELATED WITH IL5 DN | 64 | 47 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA RADIATION | 81 | 59 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| LIN MELANOMA COPY NUMBER UP | 73 | 53 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED UP | 85 | 50 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS DN | 207 | 139 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-10 | 1273 | 1279 | m8 | hsa-miR-10a | UACCCUGUAGAUCCGAAUUUGUG |
| hsa-miR-10b | UACCCUGUAGAACCGAAUUUGU | ||||
| miR-103/107 | 953 | 960 | 1A,m8 | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-129-5p | 1869 | 1875 | m8 | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-135 | 1298 | 1304 | 1A | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-153 | 608 | 614 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-221/222 | 1883 | 1890 | 1A,m8 | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-29 | 857 | 863 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-376c | 1585 | 1591 | m8 | hsa-miR-376c | AACAUAGAGGAAAUUCCACG |
| miR-448 | 608 | 614 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-450 | 1869 | 1875 | 1A | hsa-miR-450 | UUUUUGCGAUGUGUUCCUAAUA |
| miR-504 | 1274 | 1281 | 1A,m8 | hsa-miR-504 | AGACCCUGGUCUGCACUCUAU |
| miR-9 | 709 | 715 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.