Gene Page: MEIS1
Summary ?
| GeneID | 4211 |
| Symbol | MEIS1 |
| Synonyms | - |
| Description | Meis homeobox 1 |
| Reference | MIM:601739|HGNC:HGNC:7000|Ensembl:ENSG00000143995|HPRD:03442|Vega:OTTHUMG00000150714 |
| Gene type | protein-coding |
| Map location | 2p14 |
| Pascal p-value | 0.02 |
| Sherlock p-value | 0.165 |
| Fetal beta | 2.852 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 5 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06796713 | 2 | 66664871 | MEIS1 | 4.52E-6 | -0.699 | 0.01 | DMG:Wockner_2014 |
| cg06994420 | 2 | 66672553 | MEIS1 | 8.31E-6 | 0.427 | 0.013 | DMG:Wockner_2014 |
| cg03589296 | 2 | 66666292 | MEIS1 | 1.03E-4 | -0.38 | 0.028 | DMG:Wockner_2014 |
| cg00995986 | 2 | 66665428 | MEIS1 | 1.08E-4 | -0.629 | 0.028 | DMG:Wockner_2014 |
| cg12082609 | 2 | 66671727 | MEIS1 | 5.584E-4 | 0.497 | 0.049 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17392743 | chr1 | 54764240 | MEIS1 | 4211 | 0.13 | trans | ||
| rs12405921 | chr1 | 206695730 | MEIS1 | 4211 | 0.15 | trans | ||
| rs17572651 | chr1 | 218943612 | MEIS1 | 4211 | 5.194E-4 | trans | ||
| rs16829545 | chr2 | 151977407 | MEIS1 | 4211 | 9.03E-14 | trans | ||
| rs3845734 | chr2 | 171125572 | MEIS1 | 4211 | 0.02 | trans | ||
| rs7584986 | chr2 | 184111432 | MEIS1 | 4211 | 4.342E-8 | trans | ||
| rs17762315 | chr5 | 76807576 | MEIS1 | 4211 | 0.11 | trans | ||
| rs3118341 | chr9 | 25185518 | MEIS1 | 4211 | 0.03 | trans | ||
| rs11139334 | chr9 | 84209393 | MEIS1 | 4211 | 0.02 | trans | ||
| rs2393316 | chr10 | 59333070 | MEIS1 | 4211 | 5.253E-4 | trans | ||
| rs16955618 | chr15 | 29937543 | MEIS1 | 4211 | 1.143E-23 | trans | ||
| rs3790307 | chr20 | 17652571 | MEIS1 | 4211 | 0.18 | trans | ||
| rs1041786 | chr21 | 22617710 | MEIS1 | 4211 | 0.01 | trans |
Section II. Transcriptome annotation
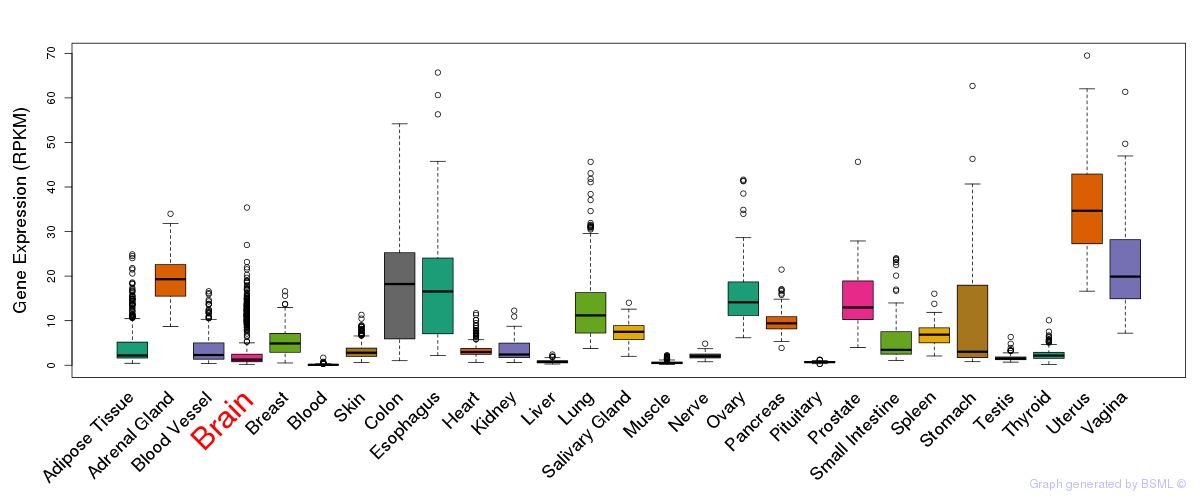
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GTPBP4 | 0.95 | 0.94 |
| IK | 0.93 | 0.95 |
| NCL | 0.93 | 0.95 |
| SNW1 | 0.93 | 0.95 |
| ZNF830 | 0.93 | 0.93 |
| HTATSF1 | 0.93 | 0.93 |
| FIP1L1 | 0.92 | 0.93 |
| OS9 | 0.92 | 0.93 |
| RIOK1 | 0.92 | 0.93 |
| CTR9 | 0.92 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.77 | -0.83 |
| MT-CO2 | -0.77 | -0.82 |
| FXYD1 | -0.76 | -0.83 |
| IFI27 | -0.75 | -0.84 |
| AF347015.33 | -0.74 | -0.80 |
| AF347015.27 | -0.73 | -0.80 |
| HIGD1B | -0.73 | -0.81 |
| HLA-F | -0.72 | -0.78 |
| MT-CYB | -0.72 | -0.78 |
| AF347015.8 | -0.71 | -0.79 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| HOXA10 | HOX1 | HOX1.8 | HOX1H | MGC12859 | PL | homeobox A10 | - | HPRD | 9343407 |
| HOXA7 | ANTP | HOX1 | HOX1.1 | HOX1A | homeobox A7 | - | HPRD | 9405651 |
| HOXA9 | ABD-B | HOX1 | HOX1.7 | HOX1G | MGC1934 | homeobox A9 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9343407 |10082572 |
| HOXB1 | HOX2 | HOX2I | Hox-2.9 | MGC116843 | MGC116844 | MGC116845 | homeobox B1 | - | HPRD,BioGRID | 10373562 |
| HOXB13 | PSGD | homeobox B13 | - | HPRD | 9343407 |
| HOXD11 | HOX4 | HOX4F | homeobox D11 | - | HPRD | 9343407 |
| HOXD12 | HOX4H | homeobox D12 | - | HPRD | 9343407 |
| HOXD9 | HOX4 | HOX4C | Hox-4.3 | Hox-5.2 | homeobox D9 | Reconstituted Complex | BioGRID | 10523646 |
| PBX1 | DKFZp686B09108 | MGC126627 | pre-B-cell leukemia homeobox 1 | Reconstituted Complex | BioGRID | 10082572 |10523646 |
| PBX2 | G17 | HOX12 | PBX2MHC | pre-B-cell leukemia homeobox 2 | - | HPRD | 10082572 |
| PBX3 | - | pre-B-cell leukemia homeobox 3 | - | HPRD | 15466398 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PYEON HPV POSITIVE TUMORS UP | 98 | 47 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| SLEBOS HEAD AND NECK CANCER WITH HPV UP | 84 | 43 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS BLUE DN | 61 | 35 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 3D UP | 182 | 110 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D UP | 157 | 91 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D UP | 194 | 122 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D UP | 175 | 108 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 UP | 276 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 2 UP | 139 | 83 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION GRANULOCYTE UP | 55 | 34 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| TOMLINS PROSTATE CANCER DN | 40 | 33 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER SOX9 TARGETS IN PROSTATE DEVELOPMENT DN | 45 | 33 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| YAO HOXA10 TARGETS VIA PROGESTERONE UP | 79 | 58 | All SZGR 2.0 genes in this pathway |
| WANG IMMORTALIZED BY HOXA9 AND MEIS1 DN | 24 | 15 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION UP | 140 | 83 | All SZGR 2.0 genes in this pathway |
| PARK HSC MARKERS | 44 | 31 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| GEISS RESPONSE TO DSRNA DN | 16 | 8 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G6 | 153 | 112 | All SZGR 2.0 genes in this pathway |
| JI RESPONSE TO FSH UP | 74 | 56 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR DN | 160 | 101 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| ROSS LEUKEMIA WITH MLL FUSIONS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 15 | 31 | 19 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION | 130 | 87 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K4ME3 AND H3K27ME3 | 38 | 34 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K4ME3 AND H327ME3 | 126 | 83 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL UP | 193 | 95 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE UP | 116 | 65 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |