Gene Page: MAP3K1
Summary ?
| GeneID | 4214 |
| Symbol | MAP3K1 |
| Synonyms | MAPKKK1|MEKK|MEKK 1|MEKK1|SRXY6 |
| Description | mitogen-activated protein kinase kinase kinase 1 |
| Reference | MIM:600982|HGNC:HGNC:6848|Ensembl:ENSG00000095015|Vega:OTTHUMG00000059486 |
| Gene type | protein-coding |
| Map location | 5q11.2 |
| Pascal p-value | 0.742 |
| Sherlock p-value | 0.853 |
| Fetal beta | 0.772 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0467 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MAP3K1 | chr5 | 56171038 | T | C | NM_005921 | . | silent | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07791897 | 5 | 55897584 | MAP3K1 | 6.39E-5 | 0.007 | 0.105 | DMG:Montano_2016 |
Section II. Transcriptome annotation
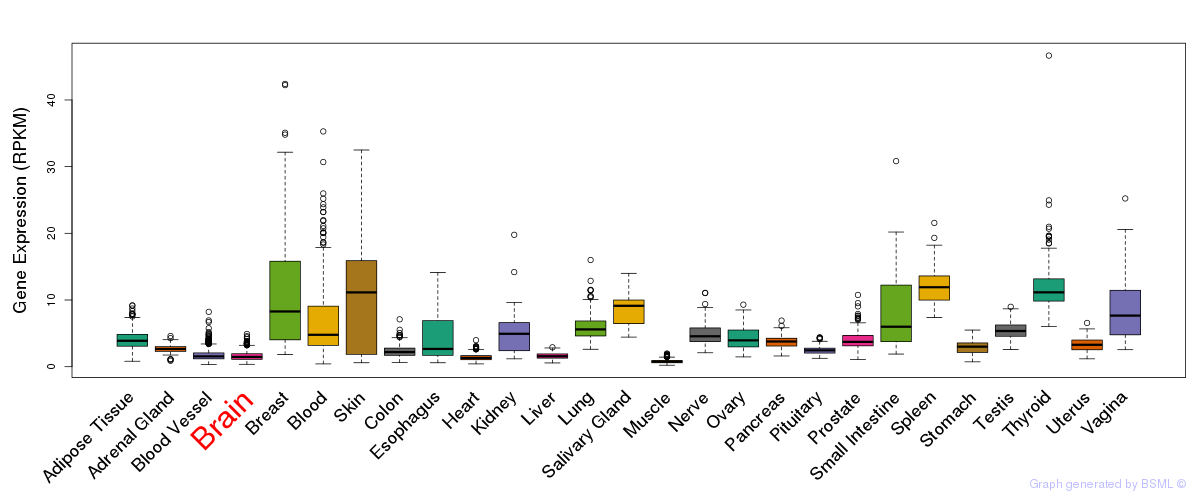
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CRB2 | 0.72 | 0.58 |
| COL4A6 | 0.70 | 0.39 |
| LRIG3 | 0.69 | 0.60 |
| NFATC4 | 0.68 | 0.54 |
| COL27A1 | 0.68 | 0.56 |
| DMRTA2 | 0.68 | 0.25 |
| DMRT3 | 0.67 | 0.41 |
| TFAP2C | 0.66 | 0.30 |
| NHLH1 | 0.66 | 0.35 |
| ITGA2 | 0.66 | 0.51 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.18 | -0.08 |
| SYCP3 | -0.17 | -0.18 |
| GNRHR | -0.17 | -0.17 |
| RP9P | -0.17 | -0.27 |
| RALYL | -0.16 | -0.16 |
| GNG8 | -0.16 | -0.20 |
| KCTD9P2 | -0.16 | -0.10 |
| AC073534.1 | -0.16 | -0.12 |
| KLHL1 | -0.16 | -0.07 |
| RTDR1 | -0.16 | -0.13 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 9808624 |11784851 |16286467 |16636664 | |
| GO:0005524 | ATP binding | NAS | - | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0004709 | MAP kinase kinase kinase activity | NAS | 9808624 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008545 | JUN kinase kinase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | NAS | 9808624 | |
| GO:0007257 | activation of JNK activity | IEA | - | |
| GO:0007179 | transforming growth factor beta receptor signaling pathway | IEA | - | |
| GO:0008637 | apoptotic mitochondrial changes | IEA | - | |
| GO:0007249 | I-kappaB kinase/NF-kappaB cascade | EXP | 15276183 | |
| GO:0042060 | wound healing | IEA | - | |
| GO:0043010 | camera-type eye development | IEA | - | |
| GO:0030838 | positive regulation of actin filament polymerization | IEA | - | |
| GO:0030334 | regulation of cell migration | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 10465784 | |
| GO:0005575 | cellular_component | ND | 9808624 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AXIN1 | AXIN | MGC52315 | axin 1 | Affinity Capture-Western | BioGRID | 10829020 |11884395 |
| CD82 | 4F9 | C33 | GR15 | IA4 | KAI1 | R2 | SAR2 | ST6 | TSPAN27 | CD82 molecule | MAP3K1 (MEKK1) interacts with the CD82 (KAI1) promoter. | BIND | 15829968 |
| CDC37 | P50CDC37 | cell division cycle 37 homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 14743216 |
| DVL2 | - | dishevelled, dsh homolog 2 (Drosophila) | Affinity Capture-Western | BioGRID | 10829020 |
| ECSIT | SITPEC | ECSIT homolog (Drosophila) | in vivo | BioGRID | 10465784 |
| FKBP5 | FKBP51 | FKBP54 | MGC111006 | P54 | PPIase | Ptg-10 | FK506 binding protein 5 | Affinity Capture-MS | BioGRID | 14743216 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9733714 |
| HSP90AA2 | HSP90ALPHA | HSPCA | HSPCAL3 | heat shock protein 90kDa alpha (cytosolic), class A member 2 | Affinity Capture-MS | BioGRID | 14743216 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | Affinity Capture-Western | BioGRID | 10969079 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | MEKK1 interacts with MEK1. | BIND | 10969079 |
| MAP2K4 | JNKK | JNKK1 | MAPKK4 | MEK4 | MKK4 | PRKMK4 | SEK1 | SERK1 | mitogen-activated protein kinase kinase 4 | MKK4 interacts with MEKK1. This interaction was modeled on a demonstrated interaction between human MKK4 and rat MEKK1. | BIND | 15866172 |
| MAP2K4 | JNKK | JNKK1 | MAPKK4 | MEK4 | MKK4 | PRKMK4 | SEK1 | SERK1 | mitogen-activated protein kinase kinase 4 | MEKK1 phosphorylates MEK4. | BIND | 10969079 |
| MAP2K7 | Jnkk2 | MAPKK7 | MKK7 | PRKMK7 | mitogen-activated protein kinase kinase 7 | MEKK1 interacts with MEK7. | BIND | 10969079 |
| MAP2K7 | Jnkk2 | MAPKK7 | MKK7 | PRKMK7 | mitogen-activated protein kinase kinase 7 | MKK7 interacts with MEKK1. This interaction was modeled on a demonstrated interaction between human MKK7 and rat MEKK1. | BIND | 15866172 |
| MAP4K2 | BL44 | GCK | RAB8IP | mitogen-activated protein kinase kinase kinase kinase 2 | Reconstituted Complex | BioGRID | 9712898 |
| MAP4K2 | BL44 | GCK | RAB8IP | mitogen-activated protein kinase kinase kinase kinase 2 | GCK interacts with, and phsophorylates, MEKK1. | BIND | 9712898 |
| MAP4K4 | FLH21957 | FLJ10410 | FLJ20373 | FLJ90111 | HGK | KIAA0687 | NIK | mitogen-activated protein kinase kinase kinase kinase 4 | Affinity Capture-Western | BioGRID | 9135144 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | Affinity Capture-Western | BioGRID | 10969079 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | MEKK1 interacts with ERK2. | BIND | 10969079 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | MEKK1 interacts with ERK1. | BIND | 12049732 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 9405400 |
| MAPK8IP3 | DKFZp762N1113 | FLJ00027 | JIP3 | JSAP1 | KIAA1066 | SYD2 | mitogen-activated protein kinase 8 interacting protein 3 | Affinity Capture-Western | BioGRID | 10523642 |
| PAK3 | CDKN1A | MRX30 | MRX47 | OPHN3 | PAK3beta | bPAK | hPAK3 | p21 protein (Cdc42/Rac)-activated kinase 3 | in vivo | BioGRID | 9065412 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | MEKK1 interacts with Raf-1. | BIND | 10969079 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10969079 |
| RPL30 | - | ribosomal protein L30 | Affinity Capture-MS | BioGRID | 14743216 |
| RPL4 | - | ribosomal protein L4 | Affinity Capture-MS | BioGRID | 14743216 |
| RPS13 | - | ribosomal protein S13 | Affinity Capture-MS | BioGRID | 14743216 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | Affinity Capture-Western Co-fractionation | BioGRID | 10346818 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Reconstituted Complex Two-hybrid | BioGRID | 9563508 |
| WDR68 | AN11 | HAN11 | WD repeat domain 68 | Affinity Capture-MS | BioGRID | 14743216 |
| YWHAE | 14-3-3E | FLJ45465 | KCIP-1 | MDCR | MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | Reconstituted Complex | BioGRID | 9452471 |