Gene Page: ATXN3
Summary ?
| GeneID | 4287 |
| Symbol | ATXN3 |
| Synonyms | AT3|ATX3|JOS|MJD|MJD1|SCA3 |
| Description | ataxin 3 |
| Reference | MIM:607047|HGNC:HGNC:7106|Ensembl:ENSG00000066427|HPRD:06131| |
| Gene type | protein-coding |
| Map location | 14q21 |
| Pascal p-value | 0.375 |
| Sherlock p-value | 0.003 |
| Fetal beta | 0.752 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0248 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg00355673 | 14 | 92573075 | ATXN3 | 8.27E-5 | -0.215 | 0.026 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs910369 | chr14 | 92530281 | ATXN3 | 4287 | 0.06 | cis | ||
| rs709930 | chr14 | 92530512 | ATXN3 | 4287 | 0.03 | cis | ||
| rs10140469 | chr14 | 92534113 | ATXN3 | 4287 | 0.06 | cis | ||
| rs8004149 | chr14 | 92534914 | ATXN3 | 4287 | 0.04 | cis | ||
| rs12588287 | chr14 | 92536958 | ATXN3 | 4287 | 0.06 | cis | ||
| rs11850101 | chr14 | 92541221 | ATXN3 | 4287 | 0.06 | cis | ||
| rs10146249 | chr14 | 92541339 | ATXN3 | 4287 | 0.11 | cis | ||
| rs1072958 | chr14 | 92552061 | ATXN3 | 4287 | 0.06 | cis | ||
| rs1072957 | chr14 | 92552071 | ATXN3 | 4287 | 0.04 | cis | ||
| rs17006218 | chr3 | 19784440 | ATXN3 | 4287 | 0.13 | trans |
Section II. Transcriptome annotation
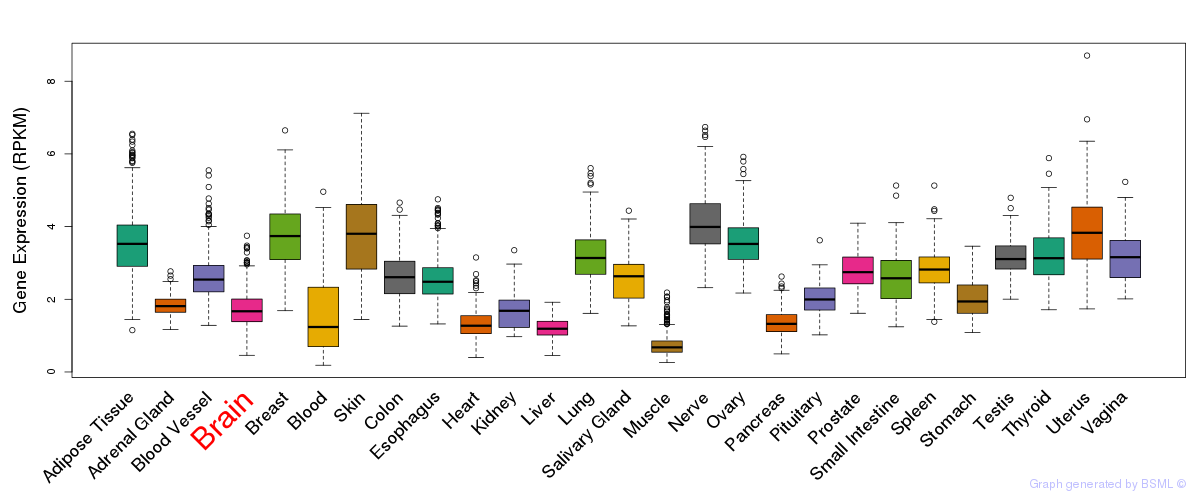
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FSTL1 | 0.78 | 0.77 |
| NID1 | 0.78 | 0.81 |
| NAGA | 0.78 | 0.73 |
| MMP14 | 0.77 | 0.73 |
| LAMA4 | 0.77 | 0.77 |
| HAPLN3 | 0.77 | 0.74 |
| HTRA3 | 0.75 | 0.76 |
| FLVCR2 | 0.75 | 0.64 |
| CXCL12 | 0.75 | 0.64 |
| STRA6 | 0.75 | 0.72 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SEPT4 | -0.42 | -0.51 |
| S100A1 | -0.42 | -0.47 |
| C5orf53 | -0.40 | -0.50 |
| AF347015.27 | -0.40 | -0.51 |
| AF347015.33 | -0.40 | -0.51 |
| HHATL | -0.39 | -0.50 |
| MT-CO2 | -0.39 | -0.51 |
| ASPA | -0.38 | -0.45 |
| AP000907.5 | -0.38 | -0.44 |
| CRYAB | -0.38 | -0.44 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 16713569 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 7655453 |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 9124802 |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006289 | nucleotide-excision repair | TAS | 10915768 | |
| GO:0008219 | cell death | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | TAS | 9580663 | |
| GO:0005737 | cytoplasm | TAS | 9124802 | |
| GO:0016363 | nuclear matrix | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO ANDROGEN UP | 29 | 21 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO FORSKOLIN DN | 9 | 7 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL DN | 101 | 66 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| KONDO EZH2 TARGETS | 245 | 148 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-125/351 | 1488 | 1494 | 1A | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-134 | 1185 | 1191 | 1A | hsa-miR-134brain | UGUGACUGGUUGACCAGAGGG |
| miR-25/32/92/363/367 | 259 | 266 | 1A,m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-9 | 190 | 196 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.