Gene Page: ACHE
Summary ?
| GeneID | 43 |
| Symbol | ACHE |
| Synonyms | ACEE|ARACHE|N-ACHE|YT |
| Description | acetylcholinesterase (Yt blood group) |
| Reference | MIM:100740|HGNC:HGNC:108|Ensembl:ENSG00000087085|HPRD:00010|Vega:OTTHUMG00000157033 |
| Gene type | protein-coding |
| Map location | 7q22 |
| Pascal p-value | 4.263E-4 |
| Sherlock p-value | 0.341 |
| Fetal beta | -1.183 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 5 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08560942 | 7 | 100492945 | ACHE | -0.016 | 0.91 | DMG:Nishioka_2013 | |
| cg27220968 | 7 | 100493724 | ACHE | 1.02E-9 | -0.013 | 1.18E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
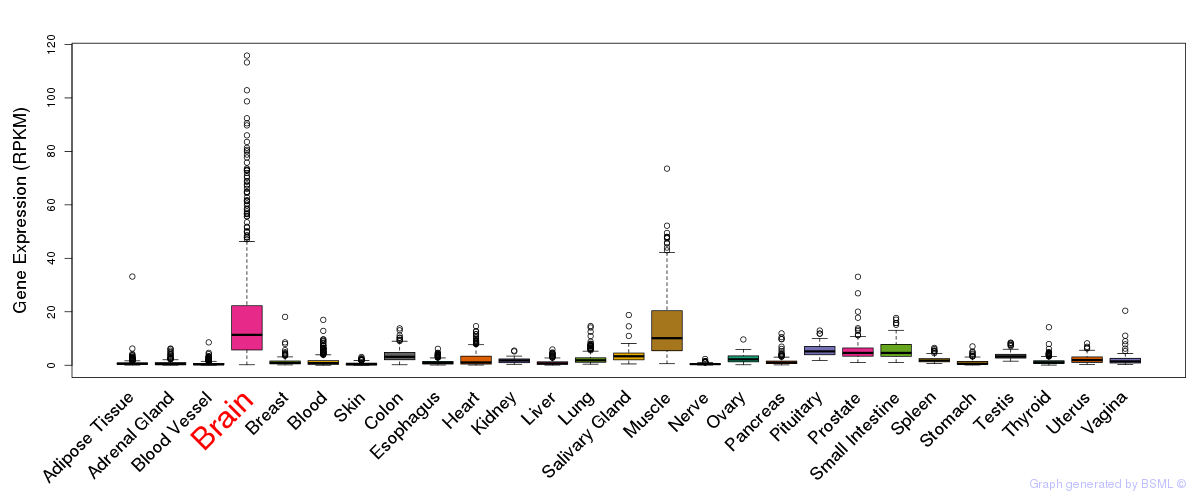
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0017171 | serine hydrolase activity | IDA | glutamate (GO term level: 4) | 3954986 |
| GO:0042166 | acetylcholine binding | NAS | Synap, Neurotransmitter (GO term level: 4) | 1517212 |
| GO:0001540 | beta-amyloid binding | TAS | 11283752 | |
| GO:0003990 | acetylcholinesterase activity | IMP | 1517212 | |
| GO:0004104 | cholinesterase activity | IDA | 1517212 | |
| GO:0004104 | cholinesterase activity | IEA | - | |
| GO:0005518 | collagen binding | IDA | 12524166 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0042803 | protein homodimerization activity | NAS | 1517212 | |
| GO:0043237 | laminin-1 binding | IDA | 12524166 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007416 | synaptogenesis | TAS | Synap (GO term level: 6) | 11283752 |
| GO:0032223 | negative regulation of synaptic transmission, cholinergic | IC | neuron, Cholinergic, Synap, Neurotransmitter (GO term level: 9) | 1517212 |
| GO:0001507 | acetylcholine catabolic process in synaptic cleft | NAS | Cholinergic, Synap (GO term level: 11) | 1517212 |
| GO:0002076 | osteoblast development | IEP | 15454088 | |
| GO:0007155 | cell adhesion | TAS | 11283752 | |
| GO:0006260 | DNA replication | TAS | 11283752 | |
| GO:0008283 | cell proliferation | TAS | 11283752 | |
| GO:0009611 | response to wounding | TAS | 11283752 | |
| GO:0007517 | muscle development | TAS | 11283752 | |
| GO:0042982 | amyloid precursor protein metabolic process | TAS | 12769797 | |
| GO:0050714 | positive regulation of protein secretion | TAS | 11283752 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045202 | synapse | IDA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | 8460160 |
| GO:0005794 | Golgi apparatus | IDA | 15454088 | |
| GO:0005576 | extracellular region | TAS | 11283752 | |
| GO:0005605 | basal lamina | NAS | 16289501 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0048471 | perinuclear region of cytoplasm | IDA | 14766237 | |
| GO:0030054 | cell junction | IEA | - | |
| GO:0031225 | anchored to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCEROPHOSPHOLIPID METABOLISM | 77 | 35 | All SZGR 2.0 genes in this pathway |
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPID METABOLISM | 198 | 112 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF PC | 18 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | 82 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| MOTAMED RESPONSE TO ANDROGEN DN | 6 | 5 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP | 207 | 128 | All SZGR 2.0 genes in this pathway |
| ROPERO HDAC2 TARGETS | 114 | 71 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER DN | 36 | 24 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE UP | 86 | 57 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT DN | 102 | 67 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| SU SALIVARY GLAND | 16 | 9 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE DN | 87 | 59 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS DN | 40 | 31 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM DN | 86 | 66 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1 TARGETS | 90 | 71 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1B TARGETS | 92 | 68 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS WITH HCP H3K27ME3 | 102 | 76 | All SZGR 2.0 genes in this pathway |
| SYED ESTRADIOL RESPONSE | 19 | 15 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-28 | 673 | 680 | 1A,m8 | hsa-miR-28brain | AAGGAGCUCACAGUCUAUUGAG |
| miR-370 | 719 | 725 | 1A | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| miR-503 | 727 | 733 | m8 | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.