Gene Page: MYOD1
Summary ?
| GeneID | 4654 |
| Symbol | MYOD1 |
| Synonyms | MYF3|MYOD|PUM|bHLHc1 |
| Description | myogenic differentiation 1 |
| Reference | MIM:159970|HGNC:HGNC:7611|Ensembl:ENSG00000129152|HPRD:01166|Vega:OTTHUMG00000166358 |
| Gene type | protein-coding |
| Map location | 11p15.4 |
| Pascal p-value | 0.558 |
| Fetal beta | -0.185 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 1.818 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24322623 | 11 | 17740430 | MYOD1 | 2.569E-4 | -0.546 | 0.038 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4757560 | chr11 | 17690161 | MYOD1 | 4654 | 0.2 | cis |
Section II. Transcriptome annotation
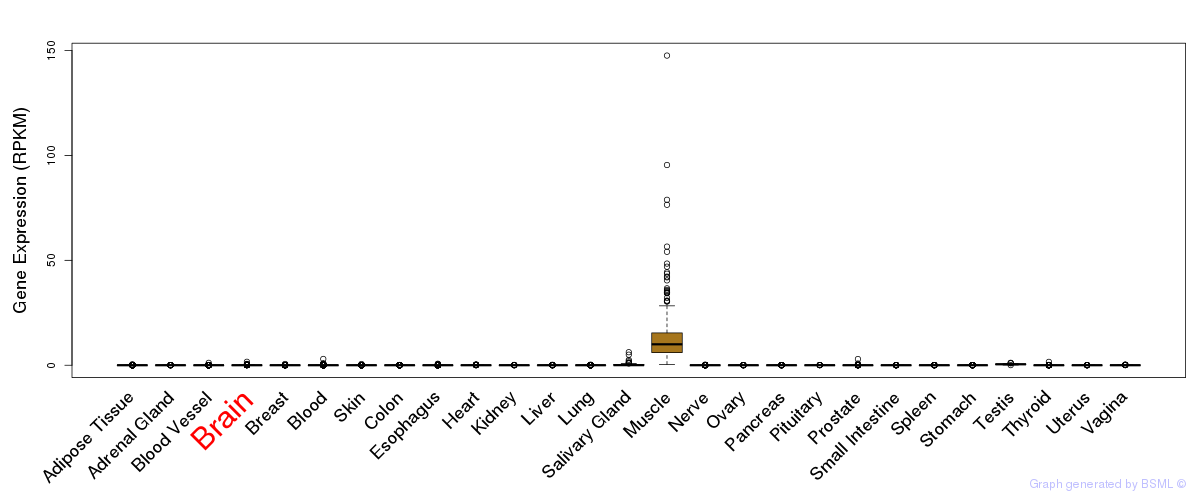
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003705 | RNA polymerase II transcription factor activity, enhancer binding | TAS | 1846704 | |
| GO:0003713 | transcription coactivator activity | TAS | 1846704 | |
| GO:0008134 | transcription factor binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007519 | skeletal muscle development | TAS | neuron (GO term level: 8) | 3175662 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | TAS | 1846704 | |
| GO:0006468 | protein amino acid phosphorylation | TAS | 3175662 | |
| GO:0007518 | myoblast cell fate determination | IEA | - | |
| GO:0007517 | muscle development | TAS | 1846704 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0045445 | myoblast differentiation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 3175662 | |
| GO:0005667 | transcription factor complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ASCL3 | HASH3 | SGN1 | bHLHa42 | achaete-scute complex homolog 3 (Drosophila) | - | HPRD | 11784080 |
| BHLHB3 | DEC2 | SHARP-1 | SHARP1 | bHLHe41 | basic helix-loop-helix domain containing, class B, 3 | - | HPRD | 12657651 |
| BHLHB8 | MIST1 | bHLHa15 | basic helix-loop-helix domain containing, class B, 8 | - | HPRD | 9482738 |
| BTG1 | - | B-cell translocation gene 1, anti-proliferative | BTG1 interacts with MYOD1 (CMD1). This interaction was modeled on a demonstrated interaction between human BTG1 and chicken CMD1. | BIND | 15674337 |
| CALM1 | CALML2 | CAMI | DD132 | PHKD | calmodulin 1 (phosphorylase kinase, delta) | - | HPRD | 10757985 |
| CALM2 | CAMII | PHKD | PHKD2 | calmodulin 2 (phosphorylase kinase, delta) | - | HPRD | 10757985 |
| CALM3 | PHKD | PHKD3 | calmodulin 3 (phosphorylase kinase, delta) | - | HPRD | 10757985 |
| CDC34 | E2-CDC34 | UBC3 | UBE2R1 | cell division cycle 34 homolog (S. cerevisiae) | - | HPRD | 9710583 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | - | HPRD,BioGRID | 10022835 |10601020 |
| CDKN1C | BWCR | BWS | KIP2 | WBS | p57 | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | - | HPRD,BioGRID | 10764802 |
| CIB2 | KIP2 | calcium and integrin binding family member 2 | - | HPRD | 10764802 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | Affinity Capture-Western Phenotypic Enhancement Reconstituted Complex | BioGRID | 9001254 |11463815 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD | 2176177 |9238849 |11463815 |
| CSRP3 | CLP | CMD1M | CMH12 | CRP3 | LMO4 | MGC14488 | MGC61993 | MLP | cysteine and glycine-rich protein 3 (cardiac LIM protein) | - | HPRD,BioGRID | 9234731 |
| ELSPBP1 | E12 | HE12 | epididymal sperm binding protein 1 | Two-hybrid | BioGRID | 9242638 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | - | HPRD,BioGRID | 9001254 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | p300 interacts with MyoD. This interaction is modelled on a demonstrated interaction between human and mouse p300 and MyoD from mouse and an unspecified species. | BIND | 8621548 |
| HAND1 | Hxt | Thing1 | bHLHa27 | eHand | heart and neural crest derivatives expressed 1 | - | HPRD | 10924525 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD,BioGRID | 11285237 |11684023 |
| HEY1 | BHLHb31 | CHF2 | HERP2 | HESR1 | HRT-1 | MGC1274 | OAF1 | hairy/enhancer-of-split related with YRPW motif 1 | - | HPRD,BioGRID | 11279181 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD,BioGRID | 1406681 |
| ID1 | ID | bHLHb24 | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein | Affinity Capture-Western Two-hybrid | BioGRID | 8380166 |9242638 |9528857 |15029197 |
| ID2 | GIG8 | ID2A | ID2H | MGC26389 | bHLHb26 | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | - | HPRD,BioGRID | 9242638 |
| ID3 | HEIR-1 | bHLHb25 | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | - | HPRD,BioGRID | 8759016 |9242638 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 1310896 |
| MDFI | I-MF | I-mfa | MyoD family inhibitor | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 8797820 |
| MEF2A | ADCAD1 | RSRFC4 | RSRFC9 | myocyte enhancer factor 2A | - | HPRD | 8548800|9418854 |
| MEF2C | - | myocyte enhancer factor 2C | Two-hybrid | BioGRID | 9418854 |
| MOS | MGC119962 | MGC119963 | MSV | v-mos Moloney murine sarcoma viral oncogene homolog | - | HPRD,BioGRID | 9001211 |
| MYOD1 | MYF3 | MYOD | PUM | bHLHc1 | myogenic differentiation 1 | Reconstituted Complex | BioGRID | 9184158 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | Reconstituted Complex | BioGRID | 10406466 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | - | HPRD | 10406466 |
| NR2F2 | ARP1 | COUP-TFII | COUPTFB | MGC117452 | SVP40 | TFCOUP2 | nuclear receptor subfamily 2, group F, member 2 | - | HPRD,BioGRID | 9826778 |
| PABPN1 | OPMD | PAB2 | PABP2 | poly(A) binding protein, nuclear 1 | Affinity Capture-Western | BioGRID | 11371506 |
| PSMD4 | AF | AF-1 | ASF | MCB1 | Rpn10 | S5A | pUB-R5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 | - | HPRD,BioGRID | 9235903 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD,BioGRID | 8381715 |
| RORA | DKFZp686M2414 | MGC119326 | MGC119329 | NR1F1 | ROR1 | ROR2 | ROR3 | RZR-ALPHA | RZRA | RAR-related orphan receptor A | - | HPRD,BioGRID | 9862959 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | - | HPRD,BioGRID | 9692544 |
| SNW1 | Bx42 | MGC119379 | NCOA-62 | PRPF45 | Prp45 | SKIIP | SKIP | SNW domain containing 1 | Affinity Capture-Western | BioGRID | 11371506 |
| SP1 | - | Sp1 transcription factor | - | HPRD,BioGRID | 10082523 |
| SRF | MCM1 | serum response factor (c-fos serum response element-binding transcription factor) | - | HPRD | 8617811 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD,BioGRID | 12947115 |
| TCF3 | E2A | ITF1 | MGC129647 | MGC129648 | bHLHb21 | transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | - | HPRD | 1649701 |9001211 |9184158 |
| TCF3 | E2A | ITF1 | MGC129647 | MGC129648 | bHLHb21 | transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 1649701 |9184158 |9242638 |
| TCF4 | E2-2 | ITF2 | MGC149723 | MGC149724 | PTHS | SEF2 | SEF2-1 | SEF2-1A | SEF2-1B | bHLHb19 | transcription factor 4 | - | HPRD,BioGRID | 8576241 |8999959 |
| TWIST1 | ACS3 | BPES2 | BPES3 | SCS | TWIST | bHLHa38 | twist homolog 1 (Drosophila) | - | HPRD | 9343420 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA HDAC PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSIII PATHWAY | 25 | 20 | All SZGR 2.0 genes in this pathway |
| PID CMYB PATHWAY | 84 | 61 | All SZGR 2.0 genes in this pathway |
| PID RB 1PATHWAY | 65 | 46 | All SZGR 2.0 genes in this pathway |
| PID HES HEY PATHWAY | 48 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME MYOGENESIS | 28 | 20 | All SZGR 2.0 genes in this pathway |
| DAVICIONI PAX FOXO1 SIGNATURE IN ARMS UP | 59 | 38 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| DAVICIONI RHABDOMYOSARCOMA PAX FOXO1 FUSION UP | 64 | 37 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| MARKS ACETYLATED NON HISTONE PROTEINS | 15 | 9 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA UP | 98 | 64 | All SZGR 2.0 genes in this pathway |
| DELASERNA MYOD TARGETS UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER UP | 27 | 20 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY OVERCONNECTED IN BREAST CANCER | 22 | 19 | All SZGR 2.0 genes in this pathway |
| YAMASHITA METHYLATED IN PROSTATE CANCER | 57 | 29 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |