Gene Page: MYT1
Summary ?
| GeneID | 4661 |
| Symbol | MYT1 |
| Synonyms | C20orf36|MTF1|MYTI|NZF2|PLPB1|ZC2HC4A |
| Description | myelin transcription factor 1 |
| Reference | MIM:600379|HGNC:HGNC:7622|Ensembl:ENSG00000196132|HPRD:02659|Vega:OTTHUMG00000149988 |
| Gene type | protein-coding |
| Map location | 20q13.33 |
| Pascal p-value | 0.242 |
| Sherlock p-value | 0.594 |
| Fetal beta | 1.537 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 1.735 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21910489 | 20 | 62818388 | MYT1 | 1.51E-5 | 0.544 | 0.015 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6687849 | chr1 | 175904808 | MYT1 | 4661 | 0.09 | trans | ||
| rs2481653 | chr1 | 176044120 | MYT1 | 4661 | 0.12 | trans | ||
| rs2502827 | chr1 | 176044216 | MYT1 | 4661 | 0 | trans | ||
| rs17572651 | chr1 | 218943612 | MYT1 | 4661 | 0.14 | trans | ||
| rs16829545 | chr2 | 151977407 | MYT1 | 4661 | 1.62E-16 | trans | ||
| rs16841750 | chr2 | 158288461 | MYT1 | 4661 | 0.06 | trans | ||
| rs3845734 | chr2 | 171125572 | MYT1 | 4661 | 1.358E-4 | trans | ||
| rs7584986 | chr2 | 184111432 | MYT1 | 4661 | 1.121E-9 | trans | ||
| rs16889813 | chr4 | 14196540 | MYT1 | 4661 | 0.17 | trans | ||
| rs2183142 | chr4 | 159232695 | MYT1 | 4661 | 0.09 | trans | ||
| rs1396222 | chr4 | 173279496 | MYT1 | 4661 | 0.15 | trans | ||
| rs335980 | chr4 | 173329784 | MYT1 | 4661 | 0.14 | trans | ||
| rs337984 | chr4 | 173411662 | MYT1 | 4661 | 0.16 | trans | ||
| rs17762315 | chr5 | 76807576 | MYT1 | 4661 | 0.02 | trans | ||
| rs16890367 | chr6 | 38078448 | MYT1 | 4661 | 0.17 | trans | ||
| rs3118341 | chr9 | 25185518 | MYT1 | 4661 | 0.01 | trans | ||
| rs11139334 | chr9 | 84209393 | MYT1 | 4661 | 0.13 | trans | ||
| rs2393316 | chr10 | 59333070 | MYT1 | 4661 | 0.11 | trans | ||
| rs17104720 | chr14 | 77127308 | MYT1 | 4661 | 0.18 | trans | ||
| rs16955618 | chr15 | 29937543 | MYT1 | 4661 | 2.458E-29 | trans | ||
| rs1041786 | chr21 | 22617710 | MYT1 | 4661 | 1.327E-5 | trans |
Section II. Transcriptome annotation
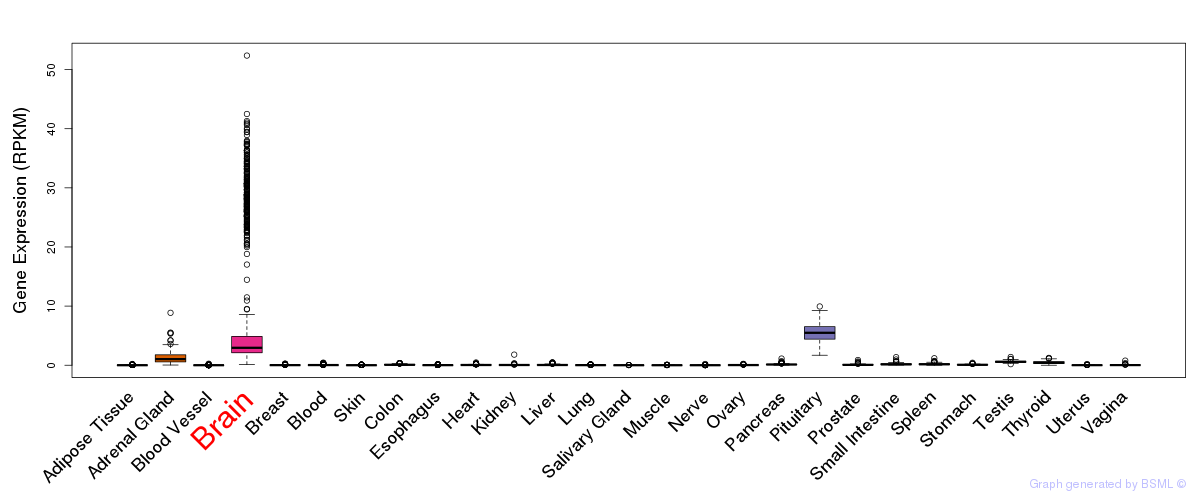
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0003700 | transcription factor activity | NAS | 1280325 | |
| GO:0005515 | protein binding | IPI | 9001210 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008270 | zinc ion binding | NAS | 1280325 | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | IEA | neurite (GO term level: 5) | - |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | NAS | 1280325 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005634 | nucleus | NAS | 1280325 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA G2 PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MPR PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RB PATHWAY | 13 | 11 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM DN | 164 | 111 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER H3K27ME3 IN NORMAL AND METHYLATED IN CANCER | 28 | 21 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 20Q12 Q13 AMPLICON | 149 | 76 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS PRENATAL | 42 | 33 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| MOLENAAR TARGETS OF CCND1 AND CDK4 UP | 67 | 48 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| ZHOU PANCREATIC ENDOCRINE PROGENITOR | 14 | 11 | All SZGR 2.0 genes in this pathway |
| VANASSE BCL2 TARGETS UP | 40 | 25 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| CARD MIR302A TARGETS | 77 | 62 | All SZGR 2.0 genes in this pathway |
| DORMOY ELAVL1 TARGETS | 17 | 10 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-125/351 | 308 | 315 | 1A,m8 | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-128 | 1708 | 1714 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-130/301 | 1052 | 1058 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-144 | 1706 | 1712 | m8 | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-146 | 305 | 311 | 1A | hsa-miR-146a | UGAGAACUGAAUUCCAUGGGUU |
| hsa-miR-146bbrain | UGAGAACUGAAUUCCAUAGGCU | ||||
| miR-186 | 1360 | 1366 | m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-200bc/429 | 336 | 342 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-27 | 1708 | 1714 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-329 | 1757 | 1763 | m8 | hsa-miR-329brain | AACACACCUGGUUAACCUCUUU |
| miR-378 | 1725 | 1731 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| miR-410 | 1342 | 1348 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-495 | 1350 | 1356 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.