Gene Page: NAP1L4
Summary ?
| GeneID | 4676 |
| Symbol | NAP1L4 |
| Synonyms | NAP1L4b|NAP2|NAP2L|hNAP2 |
| Description | nucleosome assembly protein 1 like 4 |
| Reference | MIM:601651|HGNC:HGNC:7640|Ensembl:ENSG00000205531|HPRD:11873|Vega:OTTHUMG00000011009 |
| Gene type | protein-coding |
| Map location | 11p15.5 |
| Pascal p-value | 0.553 |
| Sherlock p-value | 0.986 |
| Fetal beta | 1.214 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10318313 | 11 | 3014937 | NAP1L4 | 2.428E-4 | 0.378 | 0.037 | DMG:Wockner_2014 |
| cg04853958 | 11 | 3013544 | NAP1L4 | -0.021 | 0.36 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7104558 | chr11 | 2986775 | NAP1L4 | 4676 | 0.17 | cis |
Section II. Transcriptome annotation
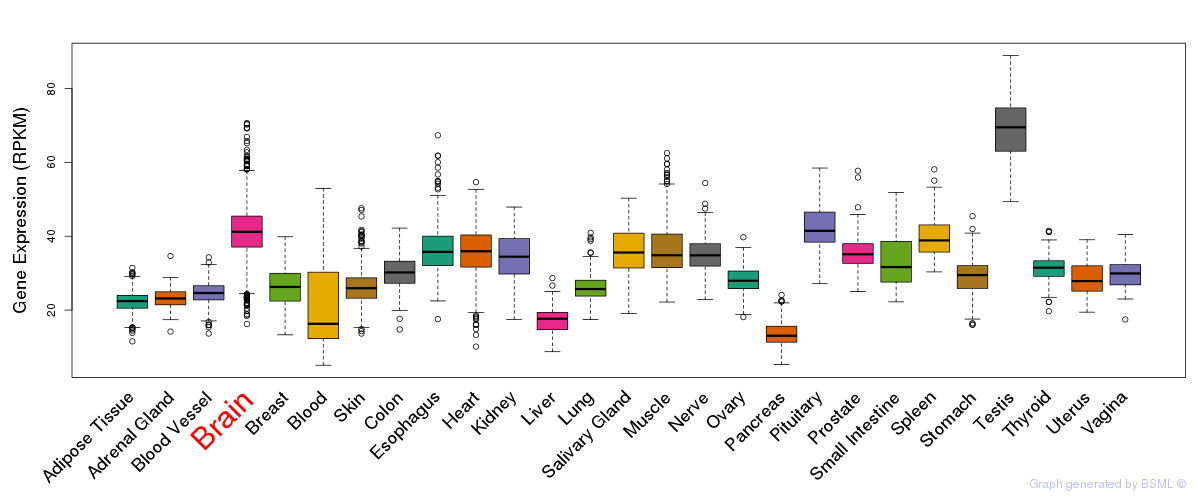
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MRPL17 | 0.88 | 0.87 |
| UROD | 0.87 | 0.86 |
| MRPS25 | 0.86 | 0.86 |
| TXNL4A | 0.86 | 0.86 |
| MECR | 0.86 | 0.85 |
| MRPL2 | 0.86 | 0.85 |
| C17orf79 | 0.86 | 0.86 |
| NDUFAB1 | 0.85 | 0.83 |
| MRPL24 | 0.85 | 0.83 |
| SNX17 | 0.85 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC010300.1 | -0.46 | -0.42 |
| AF347015.18 | -0.45 | -0.14 |
| AC005921.3 | -0.39 | -0.53 |
| C10orf108 | -0.38 | -0.29 |
| MT-ATP8 | -0.38 | -0.08 |
| EDN1 | -0.38 | -0.20 |
| EIF5B | -0.37 | -0.44 |
| AC005035.1 | -0.37 | -0.38 |
| AC004148.1 | -0.37 | -0.44 |
| AC100783.1 | -0.36 | -0.21 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| REN BOUND BY E2F | 61 | 40 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN | 128 | 90 | All SZGR 2.0 genes in this pathway |
| PARK HSC AND MULTIPOTENT PROGENITORS | 50 | 33 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 16HR | 40 | 24 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| TOOKER GEMCITABINE RESISTANCE UP | 79 | 40 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| BRIDEAU IMPRINTED GENES | 63 | 47 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |