Gene Page: NOS2
Summary ?
| GeneID | 4843 |
| Symbol | NOS2 |
| Synonyms | HEP-NOS|INOS|NOS|NOS2A |
| Description | nitric oxide synthase 2 |
| Reference | MIM:163730|HGNC:HGNC:7873|Ensembl:ENSG00000007171|HPRD:01225|Vega:OTTHUMG00000132445 |
| Gene type | protein-coding |
| Map location | 17q11.2 |
| Pascal p-value | 0.697 |
| Fetal beta | -1.748 |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NOS2 | chr17 | 26097997 | C | T | NM_000625 | p.584R>Q | missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17027208 | chr3 | 31219519 | NOS2 | 4843 | 0.11 | trans | ||
| rs1480958 | chr15 | 100575654 | NOS2 | 4843 | 0.1 | trans | ||
| rs2727185 | chr15 | 100576786 | NOS2 | 4843 | 0.12 | trans | ||
| rs4796172 | 17 | 26198381 | NOS2 | ENSG00000007171.12 | 1.246E-6 | 0.02 | -70856 | gtex_brain_ba24 |
| rs3751972 | 17 | 26206414 | NOS2 | ENSG00000007171.12 | 4.393E-6 | 0.02 | -78889 | gtex_brain_ba24 |
| rs7220420 | 17 | 26216397 | NOS2 | ENSG00000007171.12 | 3.979E-6 | 0.02 | -88872 | gtex_brain_ba24 |
| rs8078361 | 17 | 26230480 | NOS2 | ENSG00000007171.12 | 1.695E-6 | 0.02 | -102955 | gtex_brain_ba24 |
| rs7212926 | 17 | 26235226 | NOS2 | ENSG00000007171.12 | 4.666E-6 | 0.02 | -107701 | gtex_brain_ba24 |
| rs7218395 | 17 | 26241477 | NOS2 | ENSG00000007171.12 | 4.466E-6 | 0.02 | -113952 | gtex_brain_ba24 |
| rs11657699 | 17 | 26242726 | NOS2 | ENSG00000007171.12 | 1.755E-6 | 0.02 | -115201 | gtex_brain_ba24 |
| rs11657420 | 17 | 26245037 | NOS2 | ENSG00000007171.12 | 4.484E-6 | 0.02 | -117512 | gtex_brain_ba24 |
| rs1157502 | 17 | 26250109 | NOS2 | ENSG00000007171.12 | 1.695E-6 | 0.02 | -122584 | gtex_brain_ba24 |
| rs9646434 | 17 | 26251158 | NOS2 | ENSG00000007171.12 | 1.713E-6 | 0.02 | -123633 | gtex_brain_ba24 |
| rs8082467 | 17 | 26253468 | NOS2 | ENSG00000007171.12 | 1.902E-6 | 0.02 | -125943 | gtex_brain_ba24 |
| rs2531861 | 17 | 26136583 | NOS2 | ENSG00000007171.12 | 1.281E-6 | 0 | -9058 | gtex_brain_putamen_basal |
| rs2531862 | 17 | 26136991 | NOS2 | ENSG00000007171.12 | 1.317E-6 | 0 | -9466 | gtex_brain_putamen_basal |
| rs2779253 | 17 | 26137250 | NOS2 | ENSG00000007171.12 | 1.216E-6 | 0 | -9725 | gtex_brain_putamen_basal |
| rs2779254 | 17 | 26137374 | NOS2 | ENSG00000007171.12 | 1.386E-6 | 0 | -9849 | gtex_brain_putamen_basal |
| rs2779255 | 17 | 26137540 | NOS2 | ENSG00000007171.12 | 1.262E-6 | 0 | -10015 | gtex_brain_putamen_basal |
| rs2531863 | 17 | 26137550 | NOS2 | ENSG00000007171.12 | 1.265E-6 | 0 | -10025 | gtex_brain_putamen_basal |
| rs2531864 | 17 | 26138052 | NOS2 | ENSG00000007171.12 | 1.346E-6 | 0 | -10527 | gtex_brain_putamen_basal |
| rs1889022 | 17 | 26138356 | NOS2 | ENSG00000007171.12 | 1.346E-6 | 0 | -10831 | gtex_brain_putamen_basal |
| rs10853181 | 17 | 26139618 | NOS2 | ENSG00000007171.12 | 1.524E-6 | 0 | -12093 | gtex_brain_putamen_basal |
| rs2531866 | 17 | 26143164 | NOS2 | ENSG00000007171.12 | 1.524E-6 | 0 | -15639 | gtex_brain_putamen_basal |
| rs2531868 | 17 | 26143856 | NOS2 | ENSG00000007171.12 | 1.523E-6 | 0 | -16331 | gtex_brain_putamen_basal |
| rs2779257 | 17 | 26144086 | NOS2 | ENSG00000007171.12 | 1.523E-6 | 0 | -16561 | gtex_brain_putamen_basal |
| rs2531870 | 17 | 26145379 | NOS2 | ENSG00000007171.12 | 1.528E-6 | 0 | -17854 | gtex_brain_putamen_basal |
| rs2531871 | 17 | 26145437 | NOS2 | ENSG00000007171.12 | 1.528E-6 | 0 | -17912 | gtex_brain_putamen_basal |
| rs2531872 | 17 | 26147252 | NOS2 | ENSG00000007171.12 | 1.649E-6 | 0 | -19727 | gtex_brain_putamen_basal |
| rs2779258 | 17 | 26147264 | NOS2 | ENSG00000007171.12 | 7.77E-7 | 0 | -19739 | gtex_brain_putamen_basal |
| rs2531873 | 17 | 26147772 | NOS2 | ENSG00000007171.12 | 1.649E-6 | 0 | -20247 | gtex_brain_putamen_basal |
| rs2531874 | 17 | 26147797 | NOS2 | ENSG00000007171.12 | 1.649E-6 | 0 | -20272 | gtex_brain_putamen_basal |
| rs2531875 | 17 | 26148167 | NOS2 | ENSG00000007171.12 | 1.456E-6 | 0 | -20642 | gtex_brain_putamen_basal |
| rs2779259 | 17 | 26148284 | NOS2 | ENSG00000007171.12 | 1.654E-6 | 0 | -20759 | gtex_brain_putamen_basal |
| rs2531876 | 17 | 26149184 | NOS2 | ENSG00000007171.12 | 1.649E-6 | 0 | -21659 | gtex_brain_putamen_basal |
| rs2779260 | 17 | 26149327 | NOS2 | ENSG00000007171.12 | 1.649E-6 | 0 | -21802 | gtex_brain_putamen_basal |
| rs2531877 | 17 | 26149673 | NOS2 | ENSG00000007171.12 | 7.778E-7 | 0 | -22148 | gtex_brain_putamen_basal |
| rs2531878 | 17 | 26150282 | NOS2 | ENSG00000007171.12 | 6.731E-7 | 0 | -22757 | gtex_brain_putamen_basal |
| rs2531879 | 17 | 26150988 | NOS2 | ENSG00000007171.12 | 1.379E-6 | 0 | -23463 | gtex_brain_putamen_basal |
| rs2531880 | 17 | 26151021 | NOS2 | ENSG00000007171.12 | 4.969E-7 | 0 | -23496 | gtex_brain_putamen_basal |
| rs2779261 | 17 | 26151125 | NOS2 | ENSG00000007171.12 | 2.514E-6 | 0 | -23600 | gtex_brain_putamen_basal |
| rs2531881 | 17 | 26153448 | NOS2 | ENSG00000007171.12 | 1.326E-6 | 0 | -25923 | gtex_brain_putamen_basal |
| rs2779262 | 17 | 26154344 | NOS2 | ENSG00000007171.12 | 1.504E-6 | 0 | -26819 | gtex_brain_putamen_basal |
| rs2531883 | 17 | 26154725 | NOS2 | ENSG00000007171.12 | 1.318E-6 | 0 | -27200 | gtex_brain_putamen_basal |
| rs2779263 | 17 | 26155427 | NOS2 | ENSG00000007171.12 | 7.288E-7 | 0 | -27902 | gtex_brain_putamen_basal |
| rs2531886 | 17 | 26159197 | NOS2 | ENSG00000007171.12 | 1.505E-6 | 0 | -31672 | gtex_brain_putamen_basal |
| rs2531887 | 17 | 26159282 | NOS2 | ENSG00000007171.12 | 1.142E-6 | 0 | -31757 | gtex_brain_putamen_basal |
| rs2779273 | 17 | 26159786 | NOS2 | ENSG00000007171.12 | 1.437E-6 | 0 | -32261 | gtex_brain_putamen_basal |
| rs2779272 | 17 | 26161671 | NOS2 | ENSG00000007171.12 | 1.312E-6 | 0 | -34146 | gtex_brain_putamen_basal |
| rs2779271 | 17 | 26162015 | NOS2 | ENSG00000007171.12 | 1.333E-6 | 0 | -34490 | gtex_brain_putamen_basal |
| rs2779270 | 17 | 26162514 | NOS2 | ENSG00000007171.12 | 1.294E-6 | 0 | -34989 | gtex_brain_putamen_basal |
| rs2779269 | 17 | 26164000 | NOS2 | ENSG00000007171.12 | 1.337E-6 | 0 | -36475 | gtex_brain_putamen_basal |
| rs2779268 | 17 | 26164062 | NOS2 | ENSG00000007171.12 | 1.336E-6 | 0 | -36537 | gtex_brain_putamen_basal |
| rs2779267 | 17 | 26164494 | NOS2 | ENSG00000007171.12 | 1.514E-6 | 0 | -36969 | gtex_brain_putamen_basal |
| rs4063749 | 17 | 26165145 | NOS2 | ENSG00000007171.12 | 1.514E-6 | 0 | -37620 | gtex_brain_putamen_basal |
| rs139434129 | 17 | 26166971 | NOS2 | ENSG00000007171.12 | 1.883E-6 | 0 | -39446 | gtex_brain_putamen_basal |
| rs2779277 | 17 | 26170231 | NOS2 | ENSG00000007171.12 | 1.52E-6 | 0 | -42706 | gtex_brain_putamen_basal |
| rs2779278 | 17 | 26170375 | NOS2 | ENSG00000007171.12 | 1.341E-6 | 0 | -42850 | gtex_brain_putamen_basal |
| rs34350488 | 17 | 26171281 | NOS2 | ENSG00000007171.12 | 1.35E-6 | 0 | -43756 | gtex_brain_putamen_basal |
| rs4063746 | 17 | 26171381 | NOS2 | ENSG00000007171.12 | 1.343E-6 | 0 | -43856 | gtex_brain_putamen_basal |
| rs2531892 | 17 | 26173599 | NOS2 | ENSG00000007171.12 | 1.342E-6 | 0 | -46074 | gtex_brain_putamen_basal |
| rs2531893 | 17 | 26173682 | NOS2 | ENSG00000007171.12 | 1.448E-6 | 0 | -46157 | gtex_brain_putamen_basal |
| rs2531894 | 17 | 26173945 | NOS2 | ENSG00000007171.12 | 7.251E-7 | 0 | -46420 | gtex_brain_putamen_basal |
| rs2531896 | 17 | 26174102 | NOS2 | ENSG00000007171.12 | 1.3E-6 | 0 | -46577 | gtex_brain_putamen_basal |
| rs2079715 | 17 | 26177102 | NOS2 | ENSG00000007171.12 | 1.527E-8 | 0 | -49577 | gtex_brain_putamen_basal |
| rs9303698 | 17 | 26178169 | NOS2 | ENSG00000007171.12 | 1.674E-8 | 0 | -50644 | gtex_brain_putamen_basal |
| rs6505510 | 17 | 26179790 | NOS2 | ENSG00000007171.12 | 3.438E-9 | 0 | -52265 | gtex_brain_putamen_basal |
| rs9903944 | 17 | 26180739 | NOS2 | ENSG00000007171.12 | 7.822E-9 | 0 | -53214 | gtex_brain_putamen_basal |
| rs2108952 | 17 | 26181865 | NOS2 | ENSG00000007171.12 | 2.271E-10 | 0 | -54340 | gtex_brain_putamen_basal |
| rs11390208 | 17 | 26184545 | NOS2 | ENSG00000007171.12 | 2.264E-10 | 0 | -57020 | gtex_brain_putamen_basal |
| rs33943413 | 17 | 26184546 | NOS2 | ENSG00000007171.12 | 1.197E-10 | 0 | -57021 | gtex_brain_putamen_basal |
| rs7211592 | 17 | 26185418 | NOS2 | ENSG00000007171.12 | 2.278E-10 | 0 | -57893 | gtex_brain_putamen_basal |
| rs4796147 | 17 | 26186288 | NOS2 | ENSG00000007171.12 | 1.656E-10 | 0 | -58763 | gtex_brain_putamen_basal |
| rs9895004 | 17 | 26187085 | NOS2 | ENSG00000007171.12 | 2.697E-9 | 0 | -59560 | gtex_brain_putamen_basal |
| rs986993 | 17 | 26187576 | NOS2 | ENSG00000007171.12 | 1.698E-10 | 0 | -60051 | gtex_brain_putamen_basal |
| rs9674883 | 17 | 26188121 | NOS2 | ENSG00000007171.12 | 1.83E-9 | 0 | -60596 | gtex_brain_putamen_basal |
| rs28510754 | 17 | 26190828 | NOS2 | ENSG00000007171.12 | 2.767E-10 | 0 | -63303 | gtex_brain_putamen_basal |
| rs6505523 | 17 | 26192713 | NOS2 | ENSG00000007171.12 | 6.552E-9 | 0 | -65188 | gtex_brain_putamen_basal |
| rs7220473 | 17 | 26194116 | NOS2 | ENSG00000007171.12 | 5.509E-10 | 0 | -66591 | gtex_brain_putamen_basal |
| rs9909887 | 17 | 26195341 | NOS2 | ENSG00000007171.12 | 7.903E-10 | 0 | -67816 | gtex_brain_putamen_basal |
| rs12450817 | 17 | 26196802 | NOS2 | ENSG00000007171.12 | 6.206E-10 | 0 | -69277 | gtex_brain_putamen_basal |
| rs6505524 | 17 | 26197769 | NOS2 | ENSG00000007171.12 | 4.428E-10 | 0 | -70244 | gtex_brain_putamen_basal |
| rs4796172 | 17 | 26198381 | NOS2 | ENSG00000007171.12 | 2.625E-9 | 0 | -70856 | gtex_brain_putamen_basal |
| rs12936416 | 17 | 26202050 | NOS2 | ENSG00000007171.12 | 1.645E-10 | 0 | -74525 | gtex_brain_putamen_basal |
| rs1986311 | 17 | 26204098 | NOS2 | ENSG00000007171.12 | 3.523E-9 | 0 | -76573 | gtex_brain_putamen_basal |
| rs3751972 | 17 | 26206414 | NOS2 | ENSG00000007171.12 | 5.711E-10 | 0 | -78889 | gtex_brain_putamen_basal |
| rs7220420 | 17 | 26216397 | NOS2 | ENSG00000007171.12 | 3.314E-9 | 0 | -88872 | gtex_brain_putamen_basal |
| rs4796222 | 17 | 26216786 | NOS2 | ENSG00000007171.12 | 3.864E-9 | 0 | -89261 | gtex_brain_putamen_basal |
| rs2079714 | 17 | 26222520 | NOS2 | ENSG00000007171.12 | 1.909E-9 | 0 | -94995 | gtex_brain_putamen_basal |
| rs57249009 | 17 | 26222886 | NOS2 | ENSG00000007171.12 | 1.548E-9 | 0 | -95361 | gtex_brain_putamen_basal |
| rs1034895 | 17 | 26225558 | NOS2 | ENSG00000007171.12 | 7.233E-7 | 0 | -98033 | gtex_brain_putamen_basal |
| rs8078361 | 17 | 26230480 | NOS2 | ENSG00000007171.12 | 1.933E-6 | 0 | -102955 | gtex_brain_putamen_basal |
| rs2191058 | 17 | 26232917 | NOS2 | ENSG00000007171.12 | 2.369E-9 | 0 | -105392 | gtex_brain_putamen_basal |
| rs397826460 | 17 | 26235633 | NOS2 | ENSG00000007171.12 | 9.594E-8 | 0 | -108108 | gtex_brain_putamen_basal |
| rs4795142 | 17 | 26240411 | NOS2 | ENSG00000007171.12 | 5.725E-9 | 0 | -112886 | gtex_brain_putamen_basal |
| rs11657699 | 17 | 26242726 | NOS2 | ENSG00000007171.12 | 4.986E-7 | 0 | -115201 | gtex_brain_putamen_basal |
| rs9630740 | 17 | 26242756 | NOS2 | ENSG00000007171.12 | 5.952E-9 | 0 | -115231 | gtex_brain_putamen_basal |
| rs1157502 | 17 | 26250109 | NOS2 | ENSG00000007171.12 | 1.648E-6 | 0 | -122584 | gtex_brain_putamen_basal |
| rs11080380 | 17 | 26250721 | NOS2 | ENSG00000007171.12 | 3.68E-7 | 0 | -123196 | gtex_brain_putamen_basal |
| rs9646434 | 17 | 26251158 | NOS2 | ENSG00000007171.12 | 1.647E-6 | 0 | -123633 | gtex_brain_putamen_basal |
| rs11657613 | 17 | 26252856 | NOS2 | ENSG00000007171.12 | 5.953E-9 | 0 | -125331 | gtex_brain_putamen_basal |
| rs8082467 | 17 | 26253468 | NOS2 | ENSG00000007171.12 | 1.681E-6 | 0 | -125943 | gtex_brain_putamen_basal |
| rs35410059 | 17 | 26258447 | NOS2 | ENSG00000007171.12 | 5.563E-8 | 0 | -130922 | gtex_brain_putamen_basal |
| rs35652726 | 17 | 26259792 | NOS2 | ENSG00000007171.12 | 4.139E-7 | 0 | -132267 | gtex_brain_putamen_basal |
| rs55820404 | 17 | 26261503 | NOS2 | ENSG00000007171.12 | 1.458E-7 | 0 | -133978 | gtex_brain_putamen_basal |
| rs16966724 | 17 | 26262851 | NOS2 | ENSG00000007171.12 | 1.187E-6 | 0 | -135326 | gtex_brain_putamen_basal |
| rs35106758 | 17 | 26263783 | NOS2 | ENSG00000007171.12 | 2.781E-6 | 0 | -136258 | gtex_brain_putamen_basal |
| rs34371806 | 17 | 26264057 | NOS2 | ENSG00000007171.12 | 3.468E-6 | 0 | -136532 | gtex_brain_putamen_basal |
| rs34160350 | 17 | 26264319 | NOS2 | ENSG00000007171.12 | 3.473E-6 | 0 | -136794 | gtex_brain_putamen_basal |
| rs12951895 | 17 | 26264497 | NOS2 | ENSG00000007171.12 | 3.473E-6 | 0 | -136972 | gtex_brain_putamen_basal |
| rs11080045 | 17 | 26265269 | NOS2 | ENSG00000007171.12 | 5.199E-6 | 0 | -137744 | gtex_brain_putamen_basal |
| rs71372355 | 17 | 26265885 | NOS2 | ENSG00000007171.12 | 8.661E-7 | 0 | -138360 | gtex_brain_putamen_basal |
| rs12051730 | 17 | 26266230 | NOS2 | ENSG00000007171.12 | 3.473E-6 | 0 | -138705 | gtex_brain_putamen_basal |
| rs8067428 | 17 | 26267997 | NOS2 | ENSG00000007171.12 | 2.512E-6 | 0 | -140472 | gtex_brain_putamen_basal |
| rs10512428 | 17 | 26268925 | NOS2 | ENSG00000007171.12 | 2.488E-6 | 0 | -141400 | gtex_brain_putamen_basal |
| rs34256002 | 17 | 26272308 | NOS2 | ENSG00000007171.12 | 4.033E-7 | 0 | -144783 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
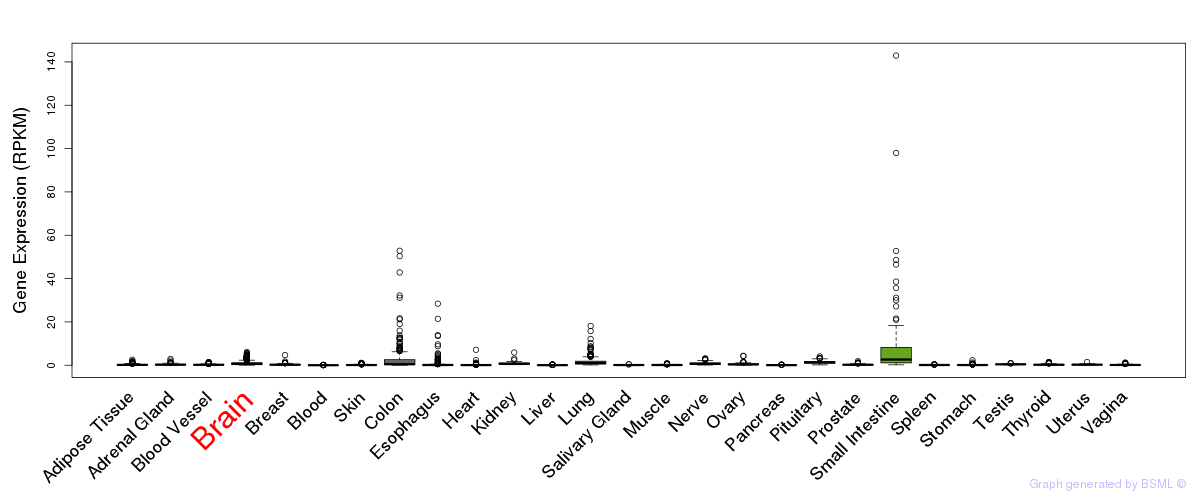
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ARGININE AND PROLINE METABOLISM | 54 | 39 | All SZGR 2.0 genes in this pathway |
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG PEROXISOME | 78 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PPARA PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NO2IL12 PATHWAY | 17 | 14 | All SZGR 2.0 genes in this pathway |
| PID IL12 2PATHWAY | 63 | 54 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A9B1 PATHWAY | 25 | 18 | All SZGR 2.0 genes in this pathway |
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| PID IL23 PATHWAY | 37 | 30 | All SZGR 2.0 genes in this pathway |
| PID P38 ALPHA BETA DOWNSTREAM PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PID HIF1 TFPATHWAY | 66 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | 33 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | 25 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET HOMEOSTASIS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL DN | 118 | 79 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| DEBOSSCHER NFKB TARGETS REPRESSED BY GLUCOCORTICOIDS | 24 | 18 | All SZGR 2.0 genes in this pathway |
| YAN ESCAPE FROM ANOIKIS | 24 | 19 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 1 | 13 | 11 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD UP | 131 | 87 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE DN | 87 | 59 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS DN | 61 | 47 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM DN | 86 | 66 | All SZGR 2.0 genes in this pathway |
| SEMENZA HIF1 TARGETS | 36 | 32 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| MIZUKAMI HYPOXIA UP | 12 | 12 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL UP | 73 | 49 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN | 214 | 124 | All SZGR 2.0 genes in this pathway |