Gene Page: PDYN
Summary ?
| GeneID | 5173 |
| Symbol | PDYN |
| Synonyms | ADCA|PENKB|SCA23 |
| Description | prodynorphin |
| Reference | MIM:131340|HGNC:HGNC:8820|Ensembl:ENSG00000101327|HPRD:08838|Vega:OTTHUMG00000031683 |
| Gene type | protein-coding |
| Map location | 20p13 |
| Pascal p-value | 0.188 |
| Sherlock p-value | 0.663 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
| Support | NEUROTROPHIN SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24377694 | 20 | 1975372 | PDYN | 1.551E-4 | 0.518 | 0.032 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6666937 | chr1 | 170286842 | PDYN | 5173 | 0.16 | trans | ||
| rs636167 | chr1 | 237402264 | PDYN | 5173 | 0.13 | trans | ||
| rs13396829 | chr2 | 120222603 | PDYN | 5173 | 0.06 | trans | ||
| rs921163 | chr2 | 205337399 | PDYN | 5173 | 0.03 | trans | ||
| rs13003798 | chr2 | 205355767 | PDYN | 5173 | 0.03 | trans | ||
| rs6746267 | chr2 | 205612604 | PDYN | 5173 | 0.16 | trans | ||
| rs16863576 | chr2 | 223120977 | PDYN | 5173 | 0.1 | trans | ||
| rs9882574 | chr3 | 56944018 | PDYN | 5173 | 0.05 | trans | ||
| rs9862967 | chr3 | 56944181 | PDYN | 5173 | 0.05 | trans | ||
| rs9825359 | chr3 | 56944297 | PDYN | 5173 | 0.05 | trans | ||
| rs9828029 | chr3 | 56949706 | PDYN | 5173 | 0.01 | trans | ||
| rs1392700 | chr3 | 56952899 | PDYN | 5173 | 0.05 | trans | ||
| rs4362744 | chr3 | 87821108 | PDYN | 5173 | 0.01 | trans | ||
| rs16827037 | chr3 | 117203284 | PDYN | 5173 | 0 | trans | ||
| rs16836473 | chr3 | 131078626 | PDYN | 5173 | 0.19 | trans | ||
| rs16847375 | chr3 | 137426961 | PDYN | 5173 | 0.17 | trans | ||
| rs17468523 | chr3 | 137427751 | PDYN | 5173 | 0.17 | trans | ||
| rs16847377 | chr3 | 137427893 | PDYN | 5173 | 0.17 | trans | ||
| rs17481423 | chr3 | 137442185 | PDYN | 5173 | 0.17 | trans | ||
| rs6834153 | chr4 | 8606484 | PDYN | 5173 | 0.01 | trans | ||
| rs7688257 | chr4 | 68126424 | PDYN | 5173 | 0.13 | trans | ||
| rs6894161 | chr5 | 91098032 | PDYN | 5173 | 0.03 | trans | ||
| rs17109939 | chr5 | 148694416 | PDYN | 5173 | 0.01 | trans | ||
| rs226963 | chr6 | 3724120 | PDYN | 5173 | 0.13 | trans | ||
| rs16885600 | chr6 | 22110853 | PDYN | 5173 | 0.1 | trans | ||
| snp_a-2046639 | 0 | PDYN | 5173 | 0.04 | trans | |||
| rs7749235 | chr6 | 30635592 | PDYN | 5173 | 0.08 | trans | ||
| rs16884095 | chr6 | 53470796 | PDYN | 5173 | 0.1 | trans | ||
| rs887719 | chr7 | 21184272 | PDYN | 5173 | 3.568E-4 | trans | ||
| rs320082 | chr7 | 29105404 | PDYN | 5173 | 2.379E-6 | trans | ||
| rs317720 | chr7 | 29110633 | PDYN | 5173 | 3.047E-4 | trans | ||
| rs317714 | chr7 | 29115553 | PDYN | 5173 | 2.17E-7 | trans | ||
| rs317724 | chr7 | 29120565 | PDYN | 5173 | 3.047E-4 | trans | ||
| rs6942437 | chr7 | 146798442 | PDYN | 5173 | 0.09 | trans | ||
| rs7829733 | chr8 | 9691720 | PDYN | 5173 | 0.06 | trans | ||
| rs10099022 | chr8 | 24510800 | PDYN | 5173 | 0.02 | trans | ||
| rs4242415 | chr8 | 24597175 | PDYN | 5173 | 0.06 | trans | ||
| rs10504043 | chr8 | 41567870 | PDYN | 5173 | 0.08 | trans | ||
| rs16886605 | chr8 | 115894398 | PDYN | 5173 | 0.04 | trans | ||
| rs7020469 | chr9 | 27338005 | PDYN | 5173 | 0.08 | trans | ||
| rs11203077 | chr10 | 91097084 | PDYN | 5173 | 0.01 | trans | ||
| rs16933545 | chr11 | 7918293 | PDYN | 5173 | 0.1 | trans | ||
| rs10834670 | chr11 | 25404015 | PDYN | 5173 | 0.12 | trans | ||
| rs11019015 | chr11 | 90157517 | PDYN | 5173 | 0.01 | trans | ||
| rs12312877 | chr12 | 79751818 | PDYN | 5173 | 0.07 | trans | ||
| rs11066476 | chr12 | 113465366 | PDYN | 5173 | 0.01 | trans | ||
| rs2993290 | chr13 | 113647189 | PDYN | 5173 | 0.01 | trans | ||
| rs1307687 | 0 | PDYN | 5173 | 0.06 | trans | |||
| rs11158850 | chr14 | 70757171 | PDYN | 5173 | 0 | trans | ||
| rs2444983 | chr15 | 33443919 | PDYN | 5173 | 0.12 | trans | ||
| rs16957838 | chr16 | 82630295 | PDYN | 5173 | 0.1 | trans | ||
| rs12452652 | chr17 | 72235560 | PDYN | 5173 | 7.708E-4 | trans | ||
| rs7230403 | chr18 | 54982203 | PDYN | 5173 | 0.18 | trans | ||
| rs1230712 | chr18 | 54992531 | PDYN | 5173 | 0.04 | trans | ||
| rs6039399 | chr20 | 9127493 | PDYN | 5173 | 0.16 | trans | ||
| rs6121844 | chr20 | 60489981 | PDYN | 5173 | 0.06 | trans |
Section II. Transcriptome annotation
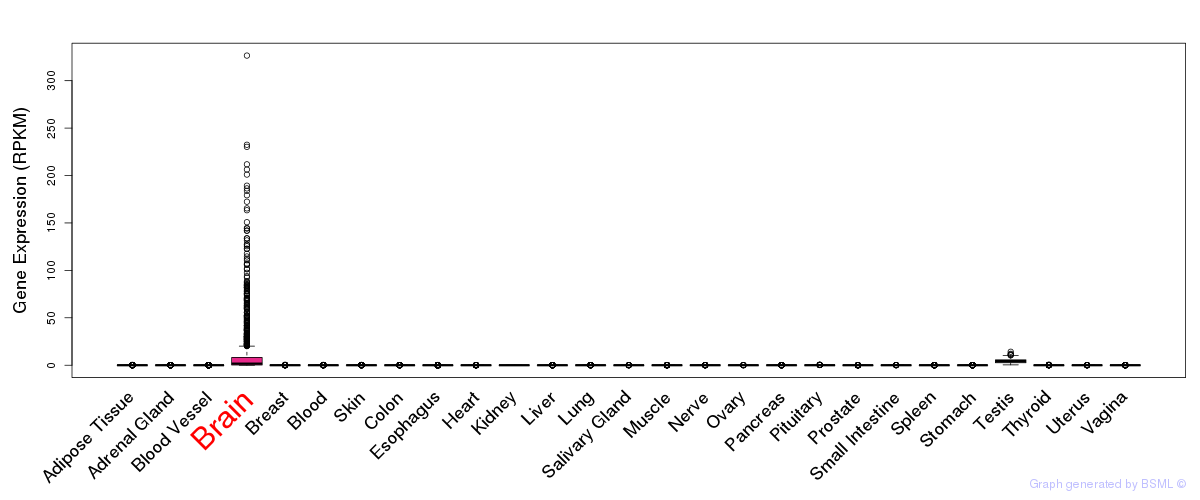
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FRMPD4 | 0.72 | 0.69 |
| THRB | 0.72 | 0.71 |
| RIMBP2 | 0.71 | 0.69 |
| ADAM22 | 0.70 | 0.69 |
| STXBP5 | 0.70 | 0.67 |
| KIF5C | 0.70 | 0.65 |
| CSMD1 | 0.69 | 0.69 |
| DLG2 | 0.69 | 0.67 |
| CYTH3 | 0.69 | 0.68 |
| POU6F2 | 0.69 | 0.64 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C1orf61 | -0.44 | -0.50 |
| SERF2 | -0.44 | -0.50 |
| WBP1 | -0.44 | -0.44 |
| ENHO | -0.43 | -0.42 |
| RPS27L | -0.43 | -0.47 |
| DBI | -0.41 | -0.43 |
| RFXANK | -0.41 | -0.46 |
| IFI27L2 | -0.40 | -0.47 |
| H2AFJ | -0.40 | -0.47 |
| TMEM219 | -0.40 | -0.40 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0001515 | opioid peptide activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | IEA | neuron, Synap, Neurotransmitter (GO term level: 6) | - |
| GO:0007218 | neuropeptide signaling pathway | IEA | Neurotransmitter (GO term level: 8) | - |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005886 | plasma membrane | IDA | 9047294 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME OPIOID SIGNALLING | 78 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME G PROTEIN ACTIVATION | 27 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE UP | 167 | 92 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND H2O2 | 39 | 29 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS DN | 57 | 39 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 2 | 50 | 34 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR UP | 71 | 48 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 2D UP | 69 | 46 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 ICP WITH H3K27ME3 | 74 | 46 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K27ME3 | 206 | 108 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K4ME3 AND H327ME3 | 126 | 83 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 AND H3K27ME3 | 137 | 85 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN UP | 56 | 38 | All SZGR 2.0 genes in this pathway |