Gene Page: PENK
Summary ?
| GeneID | 5179 |
| Symbol | PENK |
| Synonyms | PE|PENK-A |
| Description | proenkephalin |
| Reference | MIM:131330|HGNC:HGNC:8831|Ensembl:ENSG00000181195|HPRD:08837|Vega:OTTHUMG00000164409 |
| Gene type | protein-coding |
| Map location | 8q12.1 |
| Pascal p-value | 0.48 |
| Fetal beta | 0.619 |
| eGene | Cerebellar Hemisphere Myers' cis & trans |
| Support | NEUROTROPHIN SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10927978 | chr1 | 14305081 | PENK | 5179 | 0.02 | trans | ||
| rs17110698 | chr1 | 55040550 | PENK | 5179 | 0 | trans | ||
| rs596985 | chr1 | 64277034 | PENK | 5179 | 0.12 | trans | ||
| rs13447530 | chr1 | 91983798 | PENK | 5179 | 4.564E-5 | trans | ||
| rs16838771 | chr1 | 157511227 | PENK | 5179 | 4.524E-7 | trans | ||
| rs1028703 | chr1 | 167270715 | PENK | 5179 | 0.01 | trans | ||
| rs16823145 | chr1 | 184397914 | PENK | 5179 | 0.15 | trans | ||
| rs6713473 | chr2 | 11284414 | PENK | 5179 | 0.01 | trans | ||
| rs17019687 | chr2 | 80972176 | PENK | 5179 | 0.07 | trans | ||
| rs17028172 | chr2 | 103608895 | PENK | 5179 | 0.08 | trans | ||
| rs16829545 | chr2 | 151977407 | PENK | 5179 | 0.08 | trans | ||
| rs16841750 | chr2 | 158288461 | PENK | 5179 | 0.12 | trans | ||
| rs16839485 | chr2 | 208117480 | PENK | 5179 | 0.1 | trans | ||
| rs4378948 | 0 | PENK | 5179 | 0.17 | trans | |||
| rs1344868 | chr3 | 21311149 | PENK | 5179 | 0 | trans | ||
| rs4627772 | 0 | PENK | 5179 | 0.01 | trans | |||
| rs17079580 | chr3 | 47363841 | PENK | 5179 | 0.01 | trans | ||
| rs12629522 | chr3 | 59430782 | PENK | 5179 | 0.12 | trans | ||
| rs9879175 | chr3 | 59992485 | PENK | 5179 | 0.05 | trans | ||
| rs2120503 | chr3 | 66779890 | PENK | 5179 | 0.12 | trans | ||
| rs1587651 | chr3 | 79944075 | PENK | 5179 | 0.02 | trans | ||
| rs12637719 | chr3 | 85328350 | PENK | 5179 | 0.14 | trans | ||
| rs9310049 | chr3 | 87236540 | PENK | 5179 | 0.03 | trans | ||
| rs6807632 | chr3 | 111395567 | PENK | 5179 | 0.03 | trans | ||
| rs4689472 | chr4 | 6533060 | PENK | 5179 | 0.02 | trans | ||
| rs7654030 | chr4 | 65592437 | PENK | 5179 | 0.19 | trans | ||
| rs6821203 | chr4 | 189475342 | PENK | 5179 | 0.05 | trans | ||
| rs3111196 | chr5 | 54402889 | PENK | 5179 | 0.16 | trans | ||
| rs16894524 | chr5 | 65192799 | PENK | 5179 | 1.872E-4 | trans | ||
| rs17165234 | chr5 | 126403634 | PENK | 5179 | 0.07 | trans | ||
| rs1194997 | chr5 | 146093747 | PENK | 5179 | 0.11 | trans | ||
| rs6556379 | chr5 | 158399386 | PENK | 5179 | 0.01 | trans | ||
| rs17056426 | chr5 | 158452806 | PENK | 5179 | 0.12 | trans | ||
| rs16871026 | chr6 | 32815487 | PENK | 5179 | 0.2 | trans | ||
| rs794607 | chr6 | 119274656 | PENK | 5179 | 0.02 | trans | ||
| rs10499261 | chr6 | 150283162 | PENK | 5179 | 0.07 | trans | ||
| rs9397728 | chr6 | 150514248 | PENK | 5179 | 0 | trans | ||
| rs7841920 | chr8 | 74581108 | PENK | 5179 | 0.09 | trans | ||
| rs1892129 | chr9 | 21055879 | PENK | 5179 | 0.11 | trans | ||
| rs10968301 | chr9 | 28043905 | PENK | 5179 | 0.03 | trans | ||
| rs16910491 | chr9 | 116673026 | PENK | 5179 | 8.564E-4 | trans | ||
| snp_a-2108530 | 0 | PENK | 5179 | 0.01 | trans | |||
| rs16922710 | chr10 | 23087297 | PENK | 5179 | 0.13 | trans | ||
| rs16922725 | chr10 | 23100286 | PENK | 5179 | 0.17 | trans | ||
| rs10508654 | chr10 | 23104638 | PENK | 5179 | 0.05 | trans | ||
| rs7081986 | chr10 | 90807242 | PENK | 5179 | 0.13 | trans | ||
| rs4980232 | chr10 | 124577756 | PENK | 5179 | 0.03 | trans | ||
| rs2019626 | chr11 | 41926368 | PENK | 5179 | 0.12 | trans | ||
| rs12580163 | chr12 | 2134197 | PENK | 5179 | 0.08 | trans | ||
| rs11178166 | chr12 | 70670585 | PENK | 5179 | 0.18 | trans | ||
| rs17054789 | chr13 | 37561186 | PENK | 5179 | 0.11 | trans | ||
| rs17054790 | chr13 | 37562620 | PENK | 5179 | 0.11 | trans | ||
| rs336224 | chr13 | 110516305 | PENK | 5179 | 0.04 | trans | ||
| rs17104965 | chr14 | 37145915 | PENK | 5179 | 0.01 | trans | ||
| rs8013398 | chr14 | 61998391 | PENK | 5179 | 0.18 | trans | ||
| rs16955618 | chr15 | 29937543 | PENK | 5179 | 0.11 | trans | ||
| rs2118961 | chr15 | 40614199 | PENK | 5179 | 0.03 | trans | ||
| rs587388 | chr16 | 5518793 | PENK | 5179 | 0.12 | trans | ||
| rs2919431 | chr16 | 30709366 | PENK | 5179 | 0.04 | trans | ||
| rs443222 | chr16 | 83889679 | PENK | 5179 | 5.039E-4 | trans | ||
| rs824575 | chr16 | 83891459 | PENK | 5179 | 5.039E-4 | trans | ||
| rs7210845 | chr17 | 54122025 | PENK | 5179 | 2.143E-5 | trans | ||
| rs12938916 | chr17 | 55866286 | PENK | 5179 | 0.08 | trans | ||
| rs755768 | chr17 | 75023387 | PENK | 5179 | 0.09 | trans | ||
| rs11081028 | chr18 | 3269332 | PENK | 5179 | 0.05 | trans | ||
| snp_a-2036900 | 0 | PENK | 5179 | 0 | trans | |||
| rs9948702 | chr18 | 34221076 | PENK | 5179 | 0.06 | trans | ||
| rs17680225 | chr18 | 34221269 | PENK | 5179 | 0.17 | trans | ||
| rs8093472 | chr18 | 74145030 | PENK | 5179 | 0.19 | trans | ||
| rs1883720 | chr20 | 12531179 | PENK | 5179 | 0.17 | trans | ||
| rs2832308 | chr21 | 30757493 | PENK | 5179 | 9.702E-4 | trans | ||
| rs7277179 | chr21 | 30758759 | PENK | 5179 | 0.03 | trans | ||
| rs2835891 | chr21 | 39056226 | PENK | 5179 | 0.07 | trans | ||
| rs1894251 | chr22 | 22440498 | PENK | 5179 | 0.13 | trans | ||
| rs6519257 | chr22 | 41889401 | PENK | 5179 | 0.01 | trans | ||
| rs1883665 | chrX | 17650382 | PENK | 5179 | 0 | trans | ||
| rs7062071 | chrX | 17724830 | PENK | 5179 | 0.02 | trans | ||
| rs16989685 | chrX | 31527328 | PENK | 5179 | 0.11 | trans | ||
| rs5972475 | chrX | 32032409 | PENK | 5179 | 1.02E-4 | trans | ||
| rs17002014 | chrX | 138237892 | PENK | 5179 | 0.01 | trans | ||
| rs7888371 | chrX | 138238854 | PENK | 5179 | 0.01 | trans | ||
| rs7887301 | chrX | 138238881 | PENK | 5179 | 0.01 | trans |
Section II. Transcriptome annotation
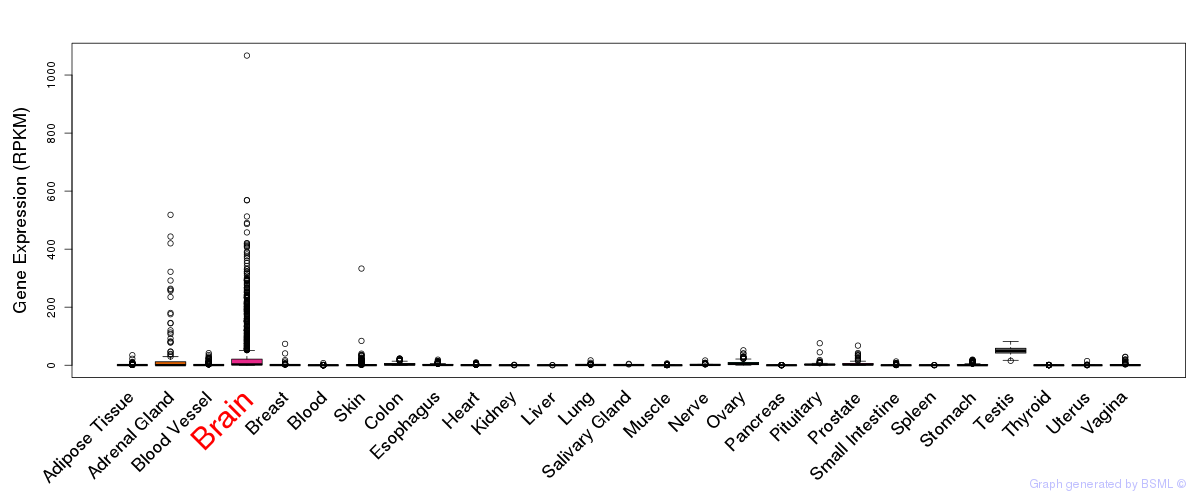
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FZR1 | 0.86 | 0.89 |
| CAPN1 | 0.86 | 0.89 |
| UBA1 | 0.85 | 0.89 |
| GRWD1 | 0.85 | 0.88 |
| PYGB | 0.85 | 0.86 |
| TBCD | 0.85 | 0.88 |
| BBS1 | 0.85 | 0.87 |
| SLC39A7 | 0.84 | 0.87 |
| RAB11B | 0.84 | 0.89 |
| TELO2 | 0.84 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.72 | -0.78 |
| MT-CO2 | -0.69 | -0.77 |
| AF347015.33 | -0.68 | -0.75 |
| AF347015.27 | -0.67 | -0.76 |
| AF347015.21 | -0.66 | -0.78 |
| C5orf53 | -0.66 | -0.68 |
| MT-CYB | -0.65 | -0.74 |
| AF347015.8 | -0.65 | -0.75 |
| IFI27 | -0.65 | -0.70 |
| AC021016.1 | -0.64 | -0.71 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005184 | neuropeptide hormone activity | TAS | axon, Synap, Brain, Neurotransmitter (GO term level: 6) | 7057924 |
| GO:0001515 | opioid peptide activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007218 | neuropeptide signaling pathway | IEA | Neurotransmitter (GO term level: 8) | - |
| GO:0001662 | behavioral fear response | IEA | - | |
| GO:0007165 | signal transduction | TAS | 7057924 | |
| GO:0007610 | behavior | IEA | - | |
| GO:0019233 | sensory perception of pain | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| CORRE MULTIPLE MYELOMA DN | 62 | 41 | All SZGR 2.0 genes in this pathway |
| LANDIS BREAST CANCER PROGRESSION DN | 70 | 43 | All SZGR 2.0 genes in this pathway |
| CASTELLANO NRAS TARGETS UP | 68 | 41 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 65 DN | 37 | 22 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST PRENEOPLASTIC DN | 55 | 33 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 DN | 149 | 93 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK UP | 116 | 68 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| HOWLIN PUBERTAL MAMMARY GLAND | 69 | 40 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS UP | 127 | 79 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR UP | 116 | 79 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER A | 100 | 63 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU DN | 83 | 69 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER UP | 49 | 36 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| TRAYNOR RETT SYNDROM UP | 45 | 33 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS DN | 57 | 39 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS UP | 37 | 27 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAINE REWARD 5D | 79 | 62 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL UP | 120 | 89 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR UP | 93 | 65 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER UP | 27 | 20 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T4 | 94 | 69 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P4 | 100 | 62 | All SZGR 2.0 genes in this pathway |
| BERNARD PPAPDC1B TARGETS UP | 40 | 20 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 16 | 26 | 17 | All SZGR 2.0 genes in this pathway |
| VALK AML WITH 11Q23 REARRANGED | 22 | 13 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION | 130 | 87 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE GENES | 90 | 54 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-200bc/429 | 185 | 192 | 1A,m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.