Gene Page: PFN2
Summary ?
| GeneID | 5217 |
| Symbol | PFN2 |
| Synonyms | D3S1319E|PFL |
| Description | profilin 2 |
| Reference | MIM:176590|HGNC:HGNC:8882|Ensembl:ENSG00000070087|HPRD:01451|Vega:OTTHUMG00000159683 |
| Gene type | protein-coding |
| Map location | 3q25.1 |
| Pascal p-value | 0.566 |
| Sherlock p-value | 0.02 |
| Fetal beta | 0.278 |
| Support | STRUCTURAL PLASTICITY CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0086 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
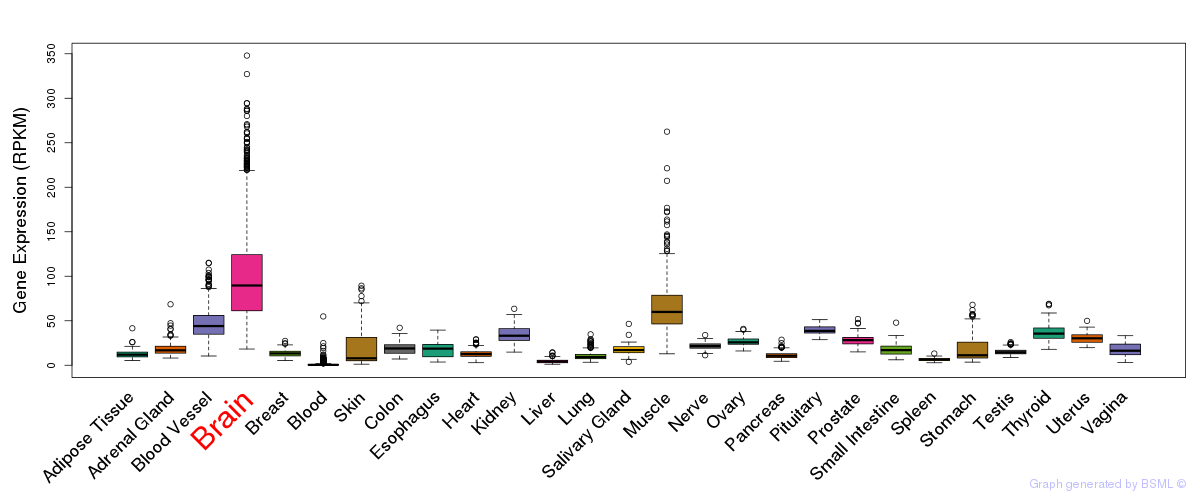
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003779 | actin binding | NAS | - | |
| GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding | NAS | - | |
| GO:0005515 | protein binding | IPI | 15383276 |16169070 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007010 | cytoskeleton organization | IEA | - | |
| GO:0008064 | regulation of actin polymerization or depolymerization | NAS | - | |
| GO:0030036 | actin cytoskeleton organization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0015629 | actin cytoskeleton | NAS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTB | PS1TP5BP1 | actin, beta | - | HPRD | 10600384 |
| ACTG1 | ACT | ACTG | DFNA20 | DFNA26 | actin, gamma 1 | Two-hybrid | BioGRID | 16169070 |
| CCDC113 | DKFZp434N1418 | FLJ23555 | HSPC065 | coiled-coil domain containing 113 | Two-hybrid | BioGRID | 16169070 |
| DDX11 | CHL1 | CHLR1 | KRG2 | MGC133249 | MGC9335 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 11 (CHL1-like helicase homolog, S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| DLST | DLTS | dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | Two-hybrid | BioGRID | 16169070 |
| DNM1 | DNM | dynamin 1 | - | HPRD | 9463375 |
| EEF1A1 | CCS-3 | CCS3 | EEF-1 | EEF1A | EF-Tu | EF1A | FLJ25721 | GRAF-1EF | HNGC:16303 | LENG7 | MGC102687 | MGC131894 | MGC16224 | PTI1 | eEF1A-1 | eukaryotic translation elongation factor 1 alpha 1 | Two-hybrid | BioGRID | 16169070 |
| EVL | RNB6 | Enah/Vasp-like | - | HPRD,BioGRID | 10945997 |
| FAM173A | C16orf24 | MGC2494 | family with sequence similarity 173, member A | Two-hybrid | BioGRID | 16169070 |
| FHOD1 | FHOS | formin homology 2 domain containing 1 | Two-hybrid | BioGRID | 12677009 |
| FHOD1 | FHOS | formin homology 2 domain containing 1 | PFN IIa interacts with FHOS. | BIND | 12677009 |
| FHOD1 | FHOS | formin homology 2 domain containing 1 | PFN IIa interacts with FHOS78. | BIND | 12677009 |
| FMNL1 | C17orf1 | C17orf1B | FHOD4 | FMNL | KW-13 | MGC133052 | MGC1894 | MGC21878 | formin-like 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10958683 |
| GOLGB1 | GCP | GCP372 | GIANTIN | GOLIM1 | golgin B1, golgi integral membrane protein | Two-hybrid | BioGRID | 16169070 |
| IK | CSA2 | MGC59741 | RED | IK cytokine, down-regulator of HLA II | Two-hybrid | BioGRID | 16169070 |
| IVNS1ABP | DKFZp686K06216 | FLARA3 | FLJ10069 | FLJ10411 | FLJ10962 | FLJ35593 | FLJ36593 | HSPC068 | KIAA0850 | ND1 | NS-1 | NS1-BP | NS1BP | influenza virus NS1A binding protein | Two-hybrid | BioGRID | 16169070 |
| LOC339344 | - | hypothetical protein LOC339344 | - | HPRD | 15615774 |
| NDUFB8 | ASHI | CI-ASHI | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19kDa | Two-hybrid | BioGRID | 16169070 |
| PLAUR | CD87 | UPAR | URKR | plasminogen activator, urokinase receptor | Two-hybrid | BioGRID | 16169070 |
| PTPRS | PTPSIGMA | protein tyrosine phosphatase, receptor type, S | Two-hybrid | BioGRID | 16169070 |
| RAP1GAP | KIAA0474 | RAP1GA1 | Rap1GAP1 | rap1GAPII | RAP1 GTPase activating protein | Two-hybrid | BioGRID | 16169070 |
| RNF146 | DKFZp434O1427 | RP3-351K20.1 | dJ351K20.1 | ring finger protein 146 | Two-hybrid | BioGRID | 16169070 |
| ROCK1 | MGC131603 | MGC43611 | P160ROCK | PRO0435 | Rho-associated, coiled-coil containing protein kinase 1 | - | HPRD,BioGRID | 14517206 |
| RPS10 | MGC88819 | ribosomal protein S10 | Two-hybrid | BioGRID | 16169070 |
| SEPT5 | CDCREL | CDCREL-1 | CDCREL1 | H5 | PNUTL1 | septin 5 | Two-hybrid | BioGRID | 16169070 |
| SGTA | SGT | alphaSGT | hSGT | small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha | Two-hybrid | BioGRID | 16169070 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | Affinity Capture-Western | BioGRID | 12391155 |
| SYN1 | SYN1a | SYN1b | SYNI | synapsin I | - | HPRD | 9463375 |
| TERF1 | FLJ41416 | PIN2 | TRBF1 | TRF | TRF1 | hTRF1-AS | t-TRF1 | telomeric repeat binding factor (NIMA-interacting) 1 | Two-hybrid | BioGRID | 16169070 |
| VASP | - | vasodilator-stimulated phosphoprotein | - | HPRD,BioGRID | 7737110 |10882740 |
| WDR33 | FLJ11294 | WDC146 | WD repeat domain 33 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | 35 | 25 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ROBO RECEPTOR | 30 | 26 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE DN | 92 | 51 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS DN | 136 | 94 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 UP | 120 | 73 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS DN | 120 | 73 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS DN | 62 | 44 | All SZGR 2.0 genes in this pathway |
| HU ANGIOGENESIS DN | 37 | 25 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| TAVOR CEBPA TARGETS DN | 30 | 21 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD DN | 162 | 102 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH REARRANGEMENTS DN | 48 | 26 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C8 | 72 | 56 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE UP | 66 | 44 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 8G | 95 | 62 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| POS RESPONSE TO HISTAMINE UP | 13 | 10 | All SZGR 2.0 genes in this pathway |
| POS HISTAMINE RESPONSE NETWORK | 32 | 22 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| CROMER TUMORIGENESIS UP | 63 | 36 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE UP | 105 | 75 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT UP | 166 | 105 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 16 | 79 | 47 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 1 UP | 125 | 61 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 2 UP | 54 | 31 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 3 UP | 170 | 97 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 4 UP | 112 | 64 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 UP | 140 | 81 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 7 UP | 118 | 68 | All SZGR 2.0 genes in this pathway |
| CARD MIR302A TARGETS | 77 | 62 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| DELPUECH FOXO3 TARGETS DN | 41 | 27 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| KRIEG KDM3A TARGETS NOT HYPOXIA | 208 | 107 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-133 | 866 | 872 | 1A | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-138 | 767 | 773 | m8 | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-150 | 401 | 407 | m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-17-5p/20/93.mr/106/519.d | 350 | 356 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU | ||||
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-183 | 959 | 966 | 1A,m8 | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
| hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG | ||||
| miR-19 | 761 | 768 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-194 | 1151 | 1157 | 1A | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-218 | 192 | 198 | 1A | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-30-5p | 1226 | 1233 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-342 | 993 | 999 | 1A | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
| miR-409-5p | 692 | 698 | 1A | hsa-miR-409-5p | AGGUUACCCGAGCAACUUUGCA |
| miR-448 | 1188 | 1194 | m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-488 | 836 | 842 | 1A | hsa-miR-488 | CCCAGAUAAUGGCACUCUCAA |
| hsa-miR-488 | CCCAGAUAAUGGCACUCUCAA | ||||
| miR-493-5p | 1093 | 1099 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-495 | 1149 | 1155 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-7 | 379 | 385 | m8 | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 349 | 355 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.