Gene Page: PHB
Summary ?
| GeneID | 5245 |
| Symbol | PHB |
| Synonyms | HEL-215|HEL-S-54e|PHB1 |
| Description | prohibitin |
| Reference | MIM:176705|HGNC:HGNC:8912|Ensembl:ENSG00000167085|HPRD:01454|Vega:OTTHUMG00000134271 |
| Gene type | protein-coding |
| Map location | 17q21 |
| Pascal p-value | 0.37 |
| Sherlock p-value | 0.047 |
| Fetal beta | -0.355 |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
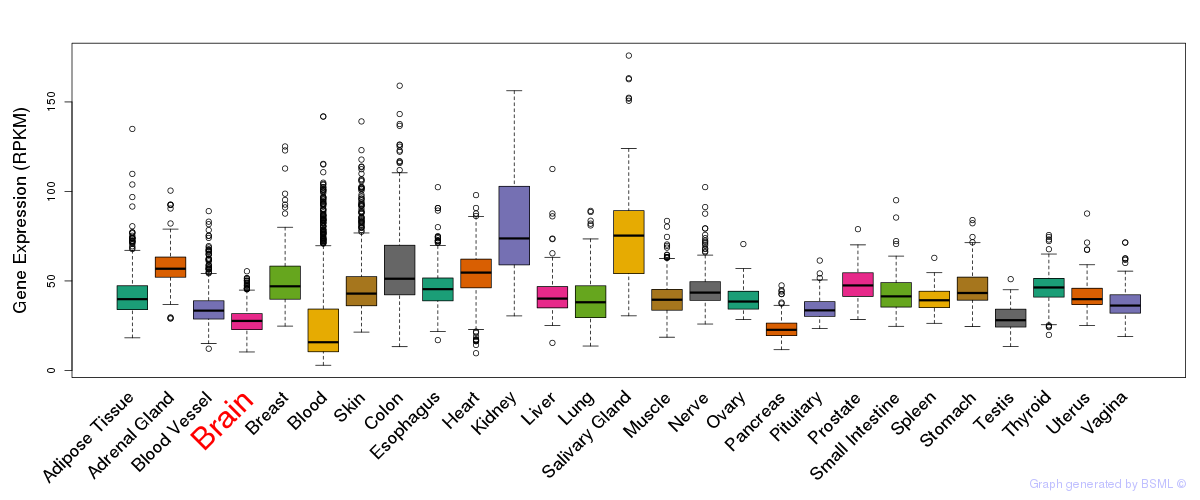
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| STAG2 | 0.86 | 0.86 |
| ZNF24 | 0.85 | 0.87 |
| POLK | 0.85 | 0.85 |
| C1orf55 | 0.84 | 0.85 |
| COPB2 | 0.84 | 0.83 |
| KIAA1826 | 0.84 | 0.85 |
| PICALM | 0.84 | 0.87 |
| EXOC5 | 0.84 | 0.84 |
| CPSF2 | 0.83 | 0.84 |
| SCARB2 | 0.83 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.65 | -0.72 |
| IFI27 | -0.64 | -0.71 |
| MT-CO2 | -0.62 | -0.68 |
| FXYD1 | -0.62 | -0.68 |
| AF347015.8 | -0.60 | -0.66 |
| CXCL14 | -0.60 | -0.68 |
| AF347015.31 | -0.59 | -0.66 |
| AF347015.27 | -0.59 | -0.64 |
| MYL3 | -0.58 | -0.63 |
| AF347015.33 | -0.58 | -0.63 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 11302691 | |
| GO:0016564 | transcription repressor activity | IDA | 12566959 | |
| GO:0016563 | transcription activator activity | IDA | 14500729 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006260 | DNA replication | IEA | - | |
| GO:0007165 | signal transduction | TAS | 16130169 | |
| GO:0008285 | negative regulation of cell proliferation | NAS | 12466959 | |
| GO:0042981 | regulation of apoptosis | TAS | 16130169 | |
| GO:0016481 | negative regulation of transcription | IDA | 12566959 | |
| GO:0016575 | histone deacetylation | IDA | 12466959 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 16130169 | |
| GO:0005654 | nucleoplasm | IDA | 12466959 | |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0005743 | mitochondrial inner membrane | IEA | - | |
| GO:0005743 | mitochondrial inner membrane | TAS | 14500729 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | IDA | 15274632 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTN1 | FLJ40884 | actinin, alpha 1 | Affinity Capture-Western | BioGRID | 12628297 |
| ANXA2 | ANX2 | ANX2L4 | CAL1H | LIP2 | LPC2 | LPC2D | P36 | PAP-IV | annexin A2 | - | HPRD,BioGRID | 12628297 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | Affinity Capture-Western Co-localization Reconstituted Complex | BioGRID | 10523633 |12065415 |14500729 |14637159 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | - | HPRD | 10523633 |14637159 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD,BioGRID | 12466959 |
| MAP3K10 | MLK2 | MST | mitogen-activated protein kinase kinase kinase 10 | - | HPRD | 9629920 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | - | HPRD,BioGRID | 12466959 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD,BioGRID | 10523633 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | RAF1 (c-Raf) interacts with PHB. | BIND | 16041367 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD,BioGRID | 10376528 |
| RBL1 | CP107 | MGC40006 | PRB1 | p107 | retinoblastoma-like 1 (p107) | in vitro in vivo | BioGRID | 10376528 |
| RBL2 | FLJ26459 | P130 | Rb2 | retinoblastoma-like 2 (p130) | - | HPRD,BioGRID | 10376528 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | Affinity Capture-Western | BioGRID | 12466959 |
| SMARCA2 | BAF190 | BRM | FLJ36757 | MGC74511 | SNF2 | SNF2L2 | SNF2LA | SWI2 | Sth1p | hBRM | hSNF2a | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | Prohibitin interacts with Brm. | BIND | 12065415 |
| SMARCA2 | BAF190 | BRM | FLJ36757 | MGC74511 | SNF2 | SNF2L2 | SNF2LA | SWI2 | Sth1p | hBRM | hSNF2a | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | - | HPRD,BioGRID | 12065415 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | - | HPRD,BioGRID | 12065415 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | Prohibitin interacts with Brg-1 | BIND | 12065415 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Affinity Capture-Western Co-localization Reconstituted Complex | BioGRID | 14500729 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA MYELOID DN | 38 | 25 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS UP | 135 | 82 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| HWANG PROSTATE CANCER MARKERS | 28 | 19 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER UP | 181 | 108 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| MENSSEN MYC TARGETS | 53 | 35 | All SZGR 2.0 genes in this pathway |
| RADAEVA RESPONSE TO IFNA1 UP | 52 | 40 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND H2O2 | 39 | 29 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 5 | 126 | 78 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN | 215 | 132 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX UP | 89 | 59 | All SZGR 2.0 genes in this pathway |
| ZHOU TNF SIGNALING 30MIN | 54 | 36 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS UP | 113 | 71 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE | 206 | 117 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS RESPONSIVE TO ESTROGEN UP | 31 | 20 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP | 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 92 | 99 | 1A,m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-205 | 768 | 774 | 1A | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG | ||||
| miR-27 | 93 | 100 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.