Gene Page: PPP1CA
Summary ?
| GeneID | 5499 |
| Symbol | PPP1CA |
| Synonyms | PP-1A|PP1A|PP1alpha|PPP1A |
| Description | protein phosphatase 1 catalytic subunit alpha |
| Reference | MIM:176875|HGNC:HGNC:9281|Ensembl:ENSG00000172531|HPRD:15942|Vega:OTTHUMG00000167671 |
| Gene type | protein-coding |
| Map location | 11q13 |
| Pascal p-value | 0.301 |
| Sherlock p-value | 0.999 |
| Fetal beta | -0.325 |
| DMG | 2 (# studies) |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 2 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.035 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg12939085 | 11 | 67166104 | PPP1CA | 2.45E-6 | 0.008 | 0.023 | DMG:Montano_2016 |
| cg11867686 | 11 | 67169368 | PPP1CA | -0.024 | 0.63 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
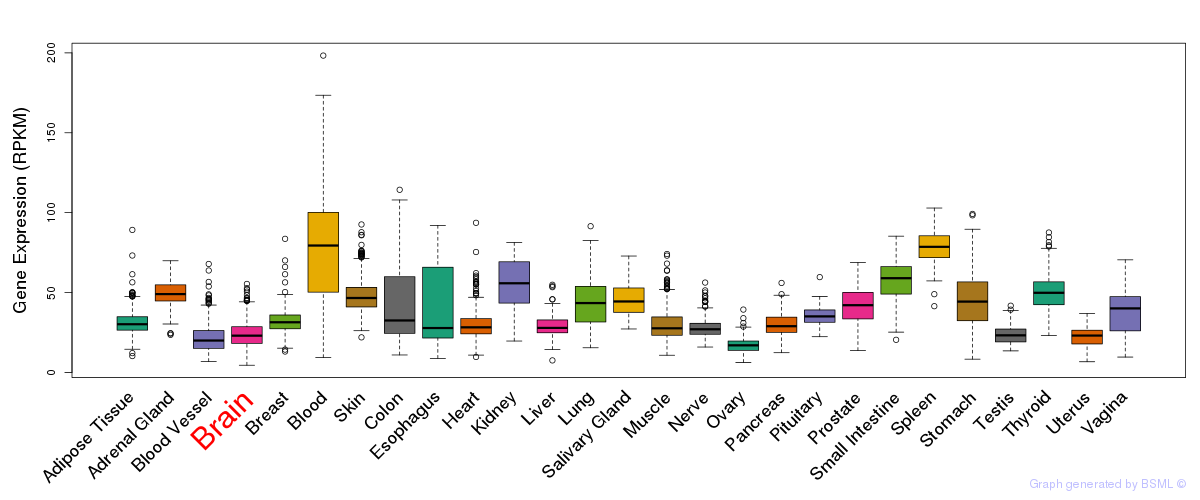
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PRMT1 | 0.97 | 0.96 |
| DDX39 | 0.95 | 0.93 |
| PAFAH1B3 | 0.95 | 0.94 |
| SMARCB1 | 0.94 | 0.92 |
| RALY | 0.94 | 0.89 |
| IP6K2 | 0.94 | 0.92 |
| ERGIC3 | 0.94 | 0.96 |
| C17orf49 | 0.94 | 0.96 |
| WRAP53 | 0.94 | 0.93 |
| NXT1 | 0.94 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.73 | -0.86 |
| AF347015.33 | -0.71 | -0.85 |
| HLA-F | -0.71 | -0.72 |
| AF347015.31 | -0.70 | -0.81 |
| MT-CO2 | -0.70 | -0.81 |
| MT-CYB | -0.69 | -0.84 |
| AF347015.8 | -0.68 | -0.84 |
| AF347015.15 | -0.68 | -0.84 |
| C5orf53 | -0.68 | -0.69 |
| AIFM3 | -0.67 | -0.69 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005506 | iron ion binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 17274640 |17511879 | |
| GO:0004722 | protein serine/threonine phosphatase activity | TAS | 8392016 | |
| GO:0004721 | phosphoprotein phosphatase activity | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0030145 | manganese ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006470 | protein amino acid dephosphorylation | IEA | - | |
| GO:0006470 | protein amino acid dephosphorylation | TAS | 8392016 | |
| GO:0005977 | glycogen metabolic process | IEA | - | |
| GO:0005975 | carbohydrate metabolic process | IEA | - | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0048754 | branching morphogenesis of a tube | IEA | - | |
| GO:0051301 | cell division | IEA | - | |
| GO:0030324 | lung development | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005654 | nucleoplasm | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTA1 | ACTA | ASMA | CFTD | CFTD1 | CFTDM | MPFD | NEM1 | NEM2 | NEM3 | actin, alpha 1, skeletal muscle | Affinity Capture-Western | BioGRID | 15107502 |
| AKAP1 | AKAP | AKAP121 | AKAP149 | AKAP84 | D-AKAP1 | MGC1807 | PRKA1 | SAKAP84 | A kinase (PRKA) anchor protein 1 | - | HPRD | 10995432 |
| AKAP11 | AKAP220 | DKFZp781I12161 | FLJ11304 | KIAA0629 | PRKA11 | A kinase (PRKA) anchor protein 11 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10209101 |12147701 |
| AKAP9 | AKAP350 | AKAP450 | CG-NAP | HYPERION | KIAA0803 | MU-RMS-40.16A | PRKA9 | YOTIAO | A kinase (PRKA) anchor protein (yotiao) 9 | - | HPRD | 10358086 |
| AURKA | AIK | ARK1 | AURA | AURORA2 | BTAK | MGC34538 | STK15 | STK6 | STK7 | aurora kinase A | - | HPRD | 11551964 |
| BAD | BBC2 | BCL2L8 | BCL2-associated agonist of cell death | - | HPRD | 10811615 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | - | HPRD | 11390485 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | - | HPRD,BioGRID | 12115603 |
| BCL2L2 | BCL-W | BCLW | KIAA0271 | BCL2-like 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12115603 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Affinity Capture-Western Reconstituted Complex | BioGRID | 12438214 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | CAV1 interacts with PP1-C. | BIND | 14645548 |
| CDC5L | CEF1 | KIAA0432 | PCDC5RP | dJ319D22.1 | hCDC5 | CDC5 cell division cycle 5-like (S. pombe) | - | HPRD | 10827081 |
| EED | HEED | WAIT1 | embryonic ectoderm development | Reconstituted Complex | BioGRID | 12788942 |
| EIF2AK2 | EIF2AK1 | MGC126524 | PKR | PRKR | eukaryotic translation initiation factor 2-alpha kinase 2 | - | HPRD,BioGRID | 12138106 |
| FXYD1 | MGC44983 | PLM | FXYD domain containing ion transport regulator 1 | - | HPRD,BioGRID | 10087003 |
| GSK3B | - | glycogen synthase kinase 3 beta | Affinity Capture-Western | BioGRID | 12147701 |
| HCFC1 | CFF | HCF-1 | HCF1 | HFC1 | MGC70925 | VCAF | host cell factor C1 (VP16-accessory protein) | Affinity Capture-Western Reconstituted Complex | BioGRID | 10637318 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | Affinity Capture-Western | BioGRID | 9269769 |
| KCNQ1 | ATFB1 | FLJ26167 | JLNS1 | KCNA8 | KCNA9 | KVLQT1 | Kv1.9 | Kv7.1 | LQT | LQT1 | RWS | SQT2 | WRS | potassium voltage-gated channel, KQT-like subfamily, member 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 11799244 |
| LMTK2 | AATYK2 | BREK | KIAA1079 | KPI-2 | KPI2 | LMR2 | cprk | lemur tyrosine kinase 2 | Affinity Capture-Western Biochemical Activity Reconstituted Complex Two-hybrid | BioGRID | 12393858 |
| MAP3K3 | MAPKKK3 | MEKK3 | mitogen-activated protein kinase kinase kinase 3 | - | HPRD | 14743216 |
| MYO16 | KIAA0865 | MYR8 | Myo16b | myosin XVI | - | HPRD | 11588169 |
| NOC2L | DKFZp564C186 | FLJ35172 | NIR | nucleolar complex associated 2 homolog (S. cerevisiae) | Reconstituted Complex | BioGRID | 10637318 |
| PHACTR1 | KIAA1733 | MGC126575 | MGC126577 | RPEL | RPEL1 | dJ257A7.2 | phosphatase and actin regulator 1 | Affinity Capture-Western Two-hybrid | BioGRID | 15107502 |
| PHACTR3 | C20orf101 | H17739 | MGC117178 | SCAPIN1 | SCAPININ | phosphatase and actin regulator 3 | - | HPRD,BioGRID | 12925532 |
| PPP1R10 | CAT53 | FB19 | PNUTS | PP1R10 | protein phosphatase 1, regulatory (inhibitor) subunit 10 | Reconstituted Complex | BioGRID | 10637318 |
| PPP1R15A | GADD34 | protein phosphatase 1, regulatory (inhibitor) subunit 15A | Affinity Capture-Western Phenotypic Suppression | BioGRID | 11564868 |12016208 |12724406 |
| PPP1R2 | IPP2 | MGC87148 | protein phosphatase 1, regulatory (inhibitor) subunit 2 | - | HPRD,BioGRID | 10807923 |
| PPP1R8 | ARD-1 | ARD1 | NIPP-1 | NIPP1 | PRO2047 | protein phosphatase 1, regulatory (inhibitor) subunit 8 | Reconstituted Complex | BioGRID | 10637318 |12788942 |
| PPP1R9A | FLJ20068 | KIAA1222 | NRB1 | NRBI | Neurabin-I | protein phosphatase 1, regulatory (inhibitor) subunit 9A | - | HPRD | 10504266 |12052877 |
| PPP1R9B | FLJ30345 | PPP1R6 | PPP1R9 | SPINO | Spn | protein phosphatase 1, regulatory (inhibitor) subunit 9B | Affinity Capture-Western Reconstituted Complex | BioGRID | 10194355 |
| RRP1B | KIAA0179 | NNP1L | Nnp1 | RRP1 | ribosomal RNA processing 1 homolog B (S. cerevisiae) | Reconstituted Complex | BioGRID | 10637318 |
| RYR2 | ARVC2 | ARVD2 | VTSIP | ryanodine receptor 2 (cardiac) | Co-fractionation | BioGRID | 10830164 |
| SMARCB1 | BAF47 | INI1 | RDT | SNF5 | SNF5L1 | Sfh1p | Snr1 | hSNFS | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 | Affinity Capture-Western Phenotypic Enhancement Reconstituted Complex | BioGRID | 12016208 |
| TGFBR1 | AAT5 | ACVRLK4 | ALK-5 | ALK5 | LDS1A | LDS2A | SKR4 | TGFR-1 | transforming growth factor, beta receptor 1 | T-beta-RI interacts with an unspecific isoform of PPP1C. This interaction was modeled on a demonstrated interaction between human T-beta-RI and mouse PPP1C. | BIND | 15761153 |
| TGFBR2 | AAT3 | FAA3 | LDS1B | LDS2B | MFS2 | RIIC | TAAD2 | TGFR-2 | TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) | T-beta-RII interacts with an unspecific isoform of PPP1C. This interaction was modeled on a demonstrated interaction between human T-beta-RII and mouse PPP1C. | BIND | 15761153 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AKAP95 PATHWAY | 12 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AKAPCENTROSOME PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CK1 PATHWAY | 17 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EIF2 PATHWAY | 11 | 8 | All SZGR 2.0 genes in this pathway |
| PID BMP PATHWAY | 42 | 31 | All SZGR 2.0 genes in this pathway |
| PID ALK1 PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PID TGFBR PATHWAY | 55 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | 23 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | 26 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME OPIOID SIGNALLING | 78 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME DARPP 32 EVENTS | 25 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | 46 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| CHOW RASSF1 TARGETS DN | 29 | 19 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| GEORGES CELL CYCLE MIR192 TARGETS | 62 | 46 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 11Q12 Q14 AMPLICON | 158 | 93 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C2 | 18 | 15 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND TBH | 60 | 42 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 | 71 | 42 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| HOFFMANN PRE BI TO LARGE PRE BII LYMPHOCYTE UP | 36 | 24 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP | 163 | 102 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| MOOTHA GLYCOGEN METABOLISM | 21 | 16 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS EARLY UP | 66 | 44 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS LATE UP | 104 | 67 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 UP | 87 | 52 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-125/351 | 236 | 242 | m8 | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-192/215 | 334 | 340 | 1A | hsa-miR-192 | CUGACCUAUGAAUUGACAGCC |
| hsa-miR-215 | AUGACCUAUGAAUUGACAGAC | ||||
| miR-335 | 323 | 329 | 1A | hsa-miR-335brain | UCAAGAGCAAUAACGAAAAAUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.