Gene Page: PPP2R1A
Summary ?
| GeneID | 5518 |
| Symbol | PPP2R1A |
| Synonyms | MRD36|PP2A-Aalpha|PP2AAALPHA|PR65A |
| Description | protein phosphatase 2 regulatory subunit A, alpha |
| Reference | MIM:605983|HGNC:HGNC:9302|HPRD:16184| |
| Gene type | protein-coding |
| Map location | 19q13.41 |
| Pascal p-value | 0.406 |
| Sherlock p-value | 0.45 |
| Fetal beta | -0.659 |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.4639 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
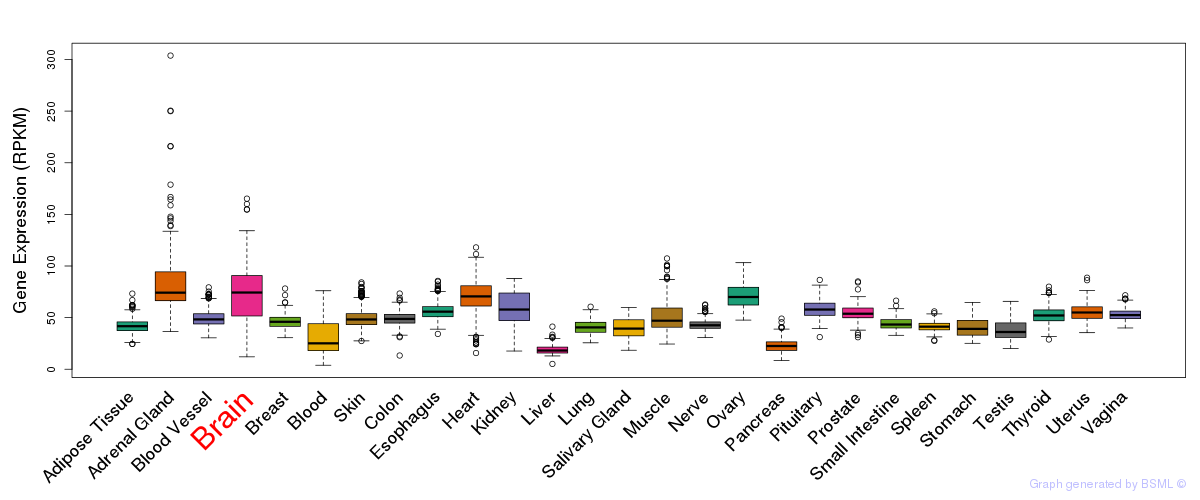
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EIF4ENIF1 | 0.92 | 0.92 |
| C6orf168 | 0.92 | 0.92 |
| TRPC4AP | 0.91 | 0.91 |
| RTN1 | 0.91 | 0.90 |
| CS | 0.91 | 0.91 |
| KIFAP3 | 0.91 | 0.92 |
| VPS26B | 0.90 | 0.91 |
| EFTUD2 | 0.90 | 0.92 |
| RUSC1 | 0.90 | 0.92 |
| AP1M1 | 0.90 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.84 | -0.86 |
| MT-CO2 | -0.83 | -0.86 |
| AF347015.8 | -0.82 | -0.87 |
| AF347015.33 | -0.82 | -0.84 |
| AF347015.27 | -0.81 | -0.85 |
| MT-CYB | -0.81 | -0.84 |
| AF347015.15 | -0.79 | -0.84 |
| AF347015.2 | -0.79 | -0.85 |
| AF347015.26 | -0.78 | -0.85 |
| FXYD1 | -0.78 | -0.83 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003823 | antigen binding | IPI | 9847399 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 9847399 | |
| GO:0008601 | protein phosphatase type 2A regulator activity | TAS | 11007961 | |
| GO:0046982 | protein heterodimerization activity | IPI | 9847399 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000188 | inactivation of MAPK activity | NAS | 11007961 | |
| GO:0008380 | RNA splicing | NAS | 11007961 | |
| GO:0006461 | protein complex assembly | TAS | 9989501 | |
| GO:0006470 | protein amino acid dephosphorylation | TAS | 11007961 | |
| GO:0010033 | response to organic substance | NAS | 11007961 | |
| GO:0006275 | regulation of DNA replication | NAS | 11007961 | |
| GO:0006917 | induction of apoptosis | TAS | 11007961 | |
| GO:0006672 | ceramide metabolic process | NAS | 11007961 | |
| GO:0030111 | regulation of Wnt receptor signaling pathway | NAS | 11007961 | |
| GO:0030155 | regulation of cell adhesion | NAS | 11007961 | |
| GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein | NAS | 11007961 | |
| GO:0040008 | regulation of growth | NAS | 11360189 | |
| GO:0019932 | second-messenger-mediated signaling | NAS | 11007961 | |
| GO:0030308 | negative regulation of cell growth | NAS | 11007961 | |
| GO:0045595 | regulation of cell differentiation | NAS | 11360189 | |
| GO:0045449 | regulation of transcription | NAS | 11007961 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000159 | protein phosphatase type 2A complex | TAS | 11007961 | |
| GO:0005829 | cytosol | IEA | - | |
| GO:0005829 | cytosol | TAS | 11007961 | |
| GO:0005625 | soluble fraction | NAS | 11007961 | |
| GO:0005634 | nucleus | NAS | 11007961 | |
| GO:0005739 | mitochondrion | NAS | 11007961 | |
| GO:0016020 | membrane | NAS | 11007961 | |
| GO:0015630 | microtubule cytoskeleton | NAS | 11007961 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARIH2 | ARI2 | FLJ10938 | FLJ33921 | TRIAD1 | ariadne homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| CTNNBIP1 | ICAT | MGC15093 | catenin, beta interacting protein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| CTTNBP2 | C7orf8 | CORTBP2 | FLJ34229 | KIAA1758 | MGC104579 | Orf4 | cortactin binding protein 2 | Affinity Capture-MS | BioGRID | 18782753 |
| CTTNBP2NL | DKFZp547A023 | FLJ13278 | CTTNBP2 N-terminal like | Affinity Capture-MS Affinity Capture-Western | BioGRID | 18782753 |
| FAM40A | FLJ14743 | KIAA1761 | MGC148091 | RP4-773N10.1 | family with sequence similarity 40, member A | Affinity Capture-MS Affinity Capture-Western | BioGRID | 18782753 |
| GNA12 | MGC104623 | MGC99644 | NNX3 | RMP | gep | guanine nucleotide binding protein (G protein) alpha 12 | Galpha12 interacts with Aalpha. This interaction was modelled on a demonstrated interaction between mouse Galpha12 and human Aalpha. | BIND | 15525651 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 10862698 |
| GRIN2D | EB11 | NMDAR2D | glutamate receptor, ionotropic, N-methyl D-aspartate 2D | - | HPRD | 10862698 |
| HSF2 | MGC117376 | MGC156196 | MGC75048 | heat shock transcription factor 2 | - | HPRD | 10224043 |
| IKBKE | IKK-i | IKKE | IKKI | KIAA0151 | MGC125294 | MGC125295 | MGC125297 | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon | Affinity Capture-MS | BioGRID | 17353931 |
| IPO9 | DKFZp761M1547 | FLJ10402 | Imp9 | importin 9 | - | HPRD | 12670497 |
| MAP3K3 | MAPKKK3 | MEKK3 | mitogen-activated protein kinase kinase kinase 3 | - | HPRD | 14743216 |
| MCC | DKFZp762O1615 | FLJ38893 | FLJ46755 | MCC1 | mutated in colorectal cancers | Affinity Capture-MS | BioGRID | 17353931 |
| MOBKL3 | 2C4D | CGI-95 | MGC12264 | MOB1 | MOB3 | PREI3 | MOB1, Mps One Binder kinase activator-like 3 (yeast) | Affinity Capture-MS | BioGRID | 18782753 |
| NFKB2 | LYT-10 | LYT10 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | - | HPRD | 14743216 |
| PDCD10 | CCM3 | MGC1212 | MGC24477 | TFAR15 | programmed cell death 10 | Affinity Capture-MS | BioGRID | 18782753 |
| PPFIA1 | FLJ41337 | FLJ42630 | FLJ43474 | LIP.1 | LIP1 | LIPRIN | MGC26800 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 | Affinity Capture-MS | BioGRID | 18782753 |
| PPFIA2 | FLJ41378 | MGC132572 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 | Affinity Capture-MS | BioGRID | 18782753 |
| PPME1 | FLJ22226 | PME-1 | protein phosphatase methylesterase 1 | Affinity Capture-MS | BioGRID | 18782753 |
| PPP2CA | PP2Ac | PP2CA | RP-C | protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 12370081 |18782753 |
| PPP2CA | PP2Ac | PP2CA | RP-C | protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform | A-alpha subunit interacts with C-alpha subunit of PP2A. | BIND | 12370081 |
| PPP2CB | PP2CB | protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 12370081 |18782753 |
| PPP2R1B | MGC26454 | PR65B | protein phosphatase 2 (formerly 2A), regulatory subunit A, beta isoform | Affinity Capture-MS | BioGRID | 18782753 |
| PPP2R2A | B55-ALPHA | B55A | FLJ26613 | MGC52248 | PR52A | PR55A | protein phosphatase 2 (formerly 2A), regulatory subunit B, alpha isoform | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 12370081 |18782753 |
| PPP2R2C | B55-GAMMA | IMYPNO | IMYPNO1 | MGC33570 | PR52 | PR55G | protein phosphatase 2 (formerly 2A), regulatory subunit B, gamma isoform | Affinity Capture-MS | BioGRID | 18782753 |
| PPP2R2D | MDS026 | protein phosphatase 2, regulatory subunit B, delta isoform | Affinity Capture-MS | BioGRID | 18782753 |
| PPP2R3B | NY-REN-8 | PPP2R3L | PPP2R3LY | PR48 | protein phosphatase 2 (formerly 2A), regulatory subunit B'', beta | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 12370081 |18782753 |
| PPP2R5A | B56A | MGC131915 | PR61A | protein phosphatase 2, regulatory subunit B', alpha isoform | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 12370081 |18782753 |
| PPP2R5C | B56G | MGC23064 | PR61G | protein phosphatase 2, regulatory subunit B', gamma isoform | Affinity Capture-MS | BioGRID | 18782753 |
| PPP2R5D | B56D | MGC2134 | MGC8949 | protein phosphatase 2, regulatory subunit B', delta isoform | Affinity Capture-MS | BioGRID | 18782753 |
| PPP2R5E | - | protein phosphatase 2, regulatory subunit B', epsilon isoform | Affinity Capture-MS | BioGRID | 18782753 |
| PPP4C | PP4 | PPH3 | PPX | protein phosphatase 4 (formerly X), catalytic subunit | Affinity Capture-MS | BioGRID | 17353931 |18715871 |
| PPP5C | FLJ36922 | PP5 | PPP5 | protein phosphatase 5, catalytic subunit | - | HPRD | 11504734 |
| RP5-1000E10.4 | DKFZp686A0768 | FLJ21168 | SIKE | suppressor of IKK epsilon | Affinity Capture-MS | BioGRID | 18782753 |
| RP6-213H19.1 | MASK | MST4 | serine/threonine protein kinase MST4 | Affinity Capture-MS | BioGRID | 18782753 |
| STK24 | MST-3 | MST3 | MST3B | STE20 | STK3 | serine/threonine kinase 24 (STE20 homolog, yeast) | Affinity Capture-MS | BioGRID | 17353931 |18782753 |
| STK25 | DKFZp686J1430 | SOK1 | YSK1 | serine/threonine kinase 25 (STE20 homolog, yeast) | Affinity Capture-MS | BioGRID | 18782753 |
| STRN | MGC125642 | SG2NA | striatin, calmodulin binding protein | Affinity Capture-MS | BioGRID | 18782753 |
| STRN3 | SG2NA | striatin, calmodulin binding protein 3 | Affinity Capture-MS | BioGRID | 18782753 |
| STRN4 | FLJ35594 | ZIN | zinedin | striatin, calmodulin binding protein 4 | Affinity Capture-MS | BioGRID | 18782753 |
| TRAF3IP3 | DJ434O14.3 | FLJ44151 | MGC117354 | MGC163289 | T3JAM | TRAF3 interacting protein 3 | Affinity Capture-MS | BioGRID | 18782753 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION | 70 | 53 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHREBP2 PATHWAY | 42 | 35 | All SZGR 2.0 genes in this pathway |
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| PID ATR PATHWAY | 39 | 25 | All SZGR 2.0 genes in this pathway |
| PID PLK1 PATHWAY | 46 | 25 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID IL8 CXCR2 PATHWAY | 34 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY WNT | 65 | 41 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME CTNNB1 PHOSPHORYLATION CASCADE | 16 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME SPRY REGULATION OF FGF SIGNALING | 14 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME TRIF MEDIATED TLR3 SIGNALING | 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | 37 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | 24 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME ERKS ARE INACTIVATED | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME ERK MAPK TARGETS | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOLYSIS | 29 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME OPIOID SIGNALLING | 78 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | 66 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME DARPP 32 EVENTS | 25 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | 59 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 PHASE | 38 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 S TRANSITION | 112 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME COSTIMULATION BY THE CD28 FAMILY | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME CTLA4 INHIBITORY SIGNALING | 21 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME MAP KINASE ACTIVATION IN TLR CASCADE | 50 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | 30 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G2 G2 M PHASES | 81 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET HOMEOSTASIS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET SENSITIZATION BY LDL | 16 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | 13 | 6 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME E2F MEDIATED REGULATION OF DNA REPLICATION | 35 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC PROMETAPHASE | 87 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE METABOLISM | 69 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | 176 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| CREIGHTON AKT1 SIGNALING VIA MTOR DN | 23 | 16 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE AUGMENTED BY MYC | 108 | 74 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| PEART HDAC PROLIFERATION CLUSTER DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS DN | 40 | 31 | All SZGR 2.0 genes in this pathway |
| KIM GASTRIC CANCER CHEMOSENSITIVITY | 103 | 64 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G4 | 21 | 15 | All SZGR 2.0 genes in this pathway |
| ZHOU TNF SIGNALING 30MIN | 54 | 36 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| URS ADIPOCYTE DIFFERENTIATION DN | 30 | 20 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND ALTERNATIVE SPLICING UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| GRADE METASTASIS DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S2 | 115 | 74 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP | 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-15/16/195/424/497 | 290 | 296 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-338 | 1 | 7 | 1A | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.