Gene Page: MYO5C
Summary ?
| GeneID | 55930 |
| Symbol | MYO5C |
| Synonyms | - |
| Description | myosin VC |
| Reference | MIM:610022|HGNC:HGNC:7604|Ensembl:ENSG00000128833|HPRD:11380|Vega:OTTHUMG00000172630 |
| Gene type | protein-coding |
| Map location | 15q21 |
| Pascal p-value | 0.146 |
| Sherlock p-value | 0.016 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07567679 | 15 | 52570612 | MIR1266;MYO5C | 2.621E-4 | 0.561 | 0.038 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17572651 | chr1 | 218943612 | MYO5C | 55930 | 0.13 | trans | ||
| rs16829545 | chr2 | 151977407 | MYO5C | 55930 | 5.986E-12 | trans | ||
| rs3845734 | chr2 | 171125572 | MYO5C | 55930 | 0 | trans | ||
| rs7584986 | chr2 | 184111432 | MYO5C | 55930 | 2.536E-7 | trans | ||
| rs2183142 | chr4 | 159232695 | MYO5C | 55930 | 0.15 | trans | ||
| rs17762315 | chr5 | 76807576 | MYO5C | 55930 | 0 | trans | ||
| rs3118341 | chr9 | 25185518 | MYO5C | 55930 | 0.03 | trans | ||
| rs17176921 | chr10 | 50580874 | MYO5C | 55930 | 0.15 | trans | ||
| rs2393316 | chr10 | 59333070 | MYO5C | 55930 | 0.05 | trans | ||
| rs16955618 | chr15 | 29937543 | MYO5C | 55930 | 2.293E-17 | trans | ||
| rs12914345 | chr15 | 77670400 | MYO5C | 55930 | 0.17 | trans | ||
| rs1041786 | chr21 | 22617710 | MYO5C | 55930 | 0 | trans | ||
| rs16986332 | chrX | 10958354 | MYO5C | 55930 | 0.04 | trans |
Section II. Transcriptome annotation
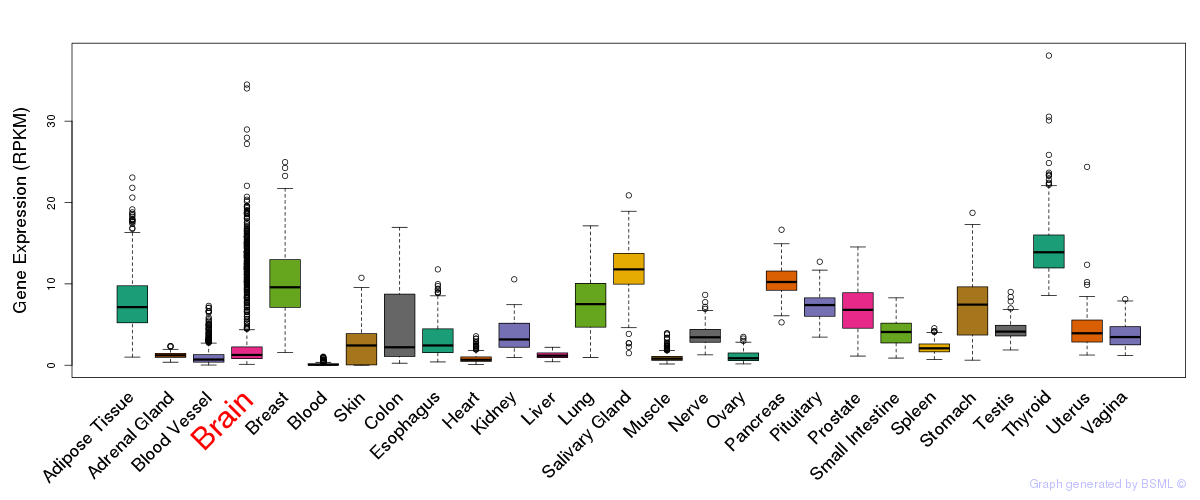
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C7orf57 | 0.98 | 0.45 |
| SPATA18 | 0.98 | 0.45 |
| YSK4 | 0.98 | 0.48 |
| ARMC3 | 0.98 | 0.56 |
| SPAG6 | 0.97 | 0.62 |
| RGS22 | 0.97 | 0.53 |
| CCDC108 | 0.97 | 0.53 |
| VWA3B | 0.97 | 0.50 |
| ARMC4 | 0.97 | 0.46 |
| TEKT1 | 0.97 | 0.44 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IER5L | -0.13 | -0.19 |
| CIDEA | -0.12 | -0.12 |
| NDRG1 | -0.12 | -0.03 |
| EMX1 | -0.11 | -0.20 |
| SLC26A4 | -0.11 | -0.09 |
| GPR22 | -0.10 | -0.15 |
| GPR21 | -0.10 | -0.07 |
| MPPED1 | -0.10 | -0.14 |
| MEF2C | -0.10 | -0.05 |
| DUSP1 | -0.10 | 0.02 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KIM RESPONSE TO TSA AND DECITABINE UP | 129 | 73 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA DN | 116 | 79 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT DN | 95 | 57 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| AIYAR COBRA1 TARGETS DN | 29 | 18 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL DN | 114 | 58 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 BULK UP | 27 | 15 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN | 169 | 102 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 3 DN | 59 | 32 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER CIPROFIBRATE UP | 60 | 42 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| HUPER BREAST BASAL VS LUMINAL DN | 59 | 44 | All SZGR 2.0 genes in this pathway |
| BEIER GLIOMA STEM CELL UP | 39 | 17 | All SZGR 2.0 genes in this pathway |
| ROSS LEUKEMIA WITH MLL FUSIONS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 UP | 167 | 99 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 11 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G1 UP | 113 | 70 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME2 | 17 | 11 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL UP | 193 | 95 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS NORMAL EMBRYOS DN | 25 | 13 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |