Gene Page: PTEN
Summary ?
| GeneID | 5728 |
| Symbol | PTEN |
| Synonyms | 10q23del|BZS|CWS1|DEC|GLM2|MHAM|MMAC1|PTEN1|TEP1 |
| Description | phosphatase and tensin homolog |
| Reference | MIM:601728|HGNC:HGNC:9588|Ensembl:ENSG00000171862|HPRD:03431|Vega:OTTHUMG00000018688 |
| Gene type | protein-coding |
| Map location | 10q23.3 |
| Pascal p-value | 0.023 |
| Sherlock p-value | 0.599 |
| Fetal beta | 0.974 |
| eGene | Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0628 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18141918 | 10 | 89622539 | PTEN;KILLIN | 1.68E-5 | -0.479 | 0.015 | DMG:Wockner_2014 |
| cg09472211 | 10 | 89622126 | PTEN;KILLIN | 1.747E-4 | -0.214 | 0.033 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16856298 | chr2 | 131650534 | PTEN | 5728 | 0.17 | trans | ||
| rs6759753 | chr2 | 131654455 | PTEN | 5728 | 0.12 | trans |
Section II. Transcriptome annotation
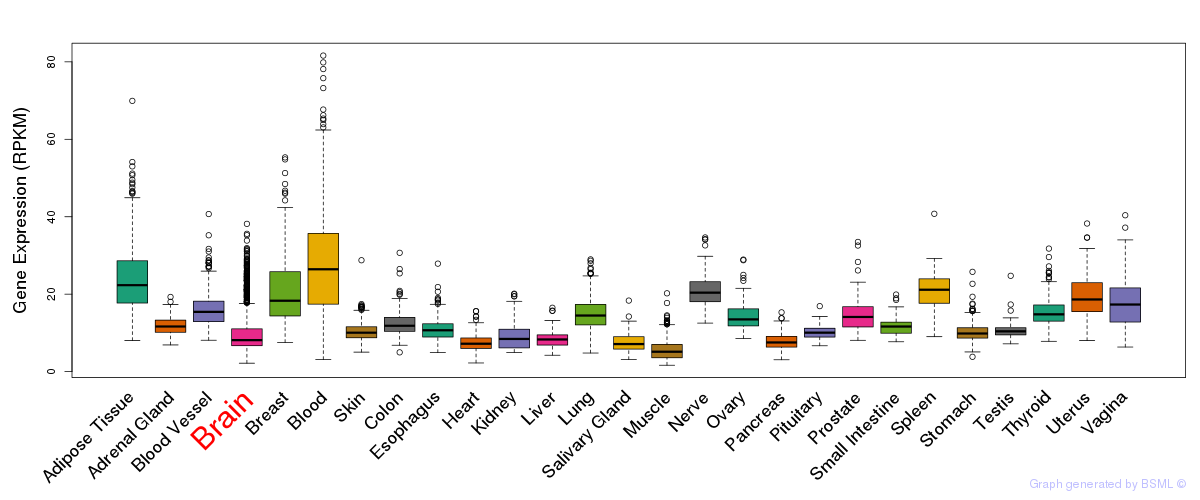
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 15951562 |17218262 |17274640 |18064632 | |
| GO:0004725 | protein tyrosine phosphatase activity | IDA | 9256433 | |
| GO:0004722 | protein serine/threonine phosphatase activity | IDA | 9256433 | |
| GO:0004438 | phosphatidylinositol-3-phosphatase activity | IDA | 9811831 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0008289 | lipid binding | IEA | - | |
| GO:0030165 | PDZ domain binding | IPI | 10646847 |10760291 | |
| GO:0008138 | protein tyrosine/serine/threonine phosphatase activity | IEA | - | |
| GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity | EXP | 9593664 | |
| GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity | IDA | 9593664 |9811831 | |
| GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity | IDA | 9593664 | |
| GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity | IDA | 9811831 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | ISS | Brain (GO term level: 6) | - |
| GO:0000079 | regulation of cyclin-dependent protein kinase activity | TAS | 10918569 | |
| GO:0001525 | angiogenesis | IEA | - | |
| GO:0006470 | protein amino acid dephosphorylation | IDA | 9256433 | |
| GO:0006470 | protein amino acid dephosphorylation | TAS | 9367992 | |
| GO:0006917 | induction of apoptosis | ISS | - | |
| GO:0008285 | negative regulation of cell proliferation | IMP | 10468583 | |
| GO:0006629 | lipid metabolic process | IEA | - | |
| GO:0043066 | negative regulation of apoptosis | IEA | - | |
| GO:0048738 | cardiac muscle development | IEA | - | |
| GO:0030336 | negative regulation of cell migration | IMP | 9616126 | |
| GO:0031647 | regulation of protein stability | IMP | 10866658 | |
| GO:0046855 | inositol phosphate dephosphorylation | IDA | 9593664 | |
| GO:0046856 | phosphoinositide dephosphorylation | IDA | 9593664 |9811831 | |
| GO:0045786 | negative regulation of cell cycle | IEA | - | |
| GO:0043542 | endothelial cell migration | IEA | - | |
| GO:0051895 | negative regulation of focal adhesion formation | IMP | 9616126 | |
| GO:0051898 | negative regulation of protein kinase B signaling cascade | IMP | 10760291 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 9593664 | |
| GO:0005737 | cytoplasm | IDA | 9187108 | |
| GO:0005737 | cytoplasm | TAS | 9367992 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANG | ALS9 | HEL168 | MGC22466 | MGC71966 | RNASE4 | RNASE5 | angiogenin, ribonuclease, RNase A family, 5 | Two-hybrid | BioGRID | 16169070 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Affinity Capture-Western Reconstituted Complex | BioGRID | 15205473 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | - | HPRD,BioGRID | 12176037 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | caveolin-1 interacts with PTEN. | BIND | 12176037 |
| CHGB | SCG1 | chromogranin B (secretogranin 1) | Two-hybrid | BioGRID | 16169070 |
| COPS6 | CSN6 | MOV34-34KD | COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis) | Two-hybrid | BioGRID | 16169070 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | - | HPRD,BioGRID | 12297295 |
| CSNK2A2 | CK2A2 | CSNK2A1 | FLJ43934 | casein kinase 2, alpha prime polypeptide | Biochemical Activity Reconstituted Complex | BioGRID | 12297295 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | Reconstituted Complex | BioGRID | 15205473 |
| GLTSCR2 | PICT-1 | PICT1 | glioma tumor suppressor candidate region gene 2 | PTEN interacts with PICT-1. | BIND | 15355975 |
| HBA1 | CD31 | MGC126895 | MGC126897 | hemoglobin, alpha 1 | Two-hybrid | BioGRID | 16169070 |
| MAGI2 | ACVRIP1 | AIP1 | ARIP1 | MAGI-2 | SSCAM | membrane associated guanylate kinase, WW and PDZ domain containing 2 | - | HPRD | 10760291 |
| MAGI3 | MAGI-3 | MGC163281 | RP4-730K3.1 | dJ730K3.2 | membrane associated guanylate kinase, WW and PDZ domain containing 3 | - | HPRD,BioGRID | 10748157 |
| MAGI3 | MAGI-3 | MGC163281 | RP4-730K3.1 | dJ730K3.2 | membrane associated guanylate kinase, WW and PDZ domain containing 3 | MAGI3 interacts with PTEN/MMAC. | BIND | 10748157 |
| MVP | LRP | VAULT1 | major vault protein | - | HPRD,BioGRID | 12177006 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD,BioGRID | 10400703 |
| PXN | FLJ16691 | paxillin | - | HPRD,BioGRID | 11857088 |
| ROCK1 | MGC131603 | MGC43611 | P160ROCK | PRO0435 | Rho-associated, coiled-coil containing protein kinase 1 | PTEN interacts with Rock1. | BIND | 15793569 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 12620407 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Affinity Capture-Western | BioGRID | 12620973 |
| UBE2L3 | E2-F1 | L-UBC | UBCH7 | UbcM4 | ubiquitin-conjugating enzyme E2L 3 | - | HPRD,BioGRID | 12620973 |
| UTP14A | KIAA0266 | NY-CO-16 | SDCCAG16 | dJ537K23.3 | UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG INOSITOL PHOSPHATE METABOLISM | 54 | 42 | All SZGR 2.0 genes in this pathway |
| KEGG PHOSPHATIDYLINOSITOL SIGNALING SYSTEM | 76 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOMETRIAL CANCER | 52 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOMA | 71 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CTCF PATHWAY | 23 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MTOR PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PTEN PATHWAY | 18 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EIF4 PATHWAY | 24 | 19 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MET PATHWAY | 37 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1MTOR PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| SIG CHEMOTAXIS | 45 | 37 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | 51 | 41 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN B LYMPHOCYTES | 36 | 31 | All SZGR 2.0 genes in this pathway |
| SA PTEN PATHWAY | 17 | 14 | All SZGR 2.0 genes in this pathway |
| ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| PID RHOA PATHWAY | 45 | 33 | All SZGR 2.0 genes in this pathway |
| PID TCR PATHWAY | 66 | 51 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI PATHWAY | 49 | 40 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID KIT PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY SCF KIT | 78 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB4 SIGNALING | 38 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB2 SIGNALING | 44 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | 97 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPID METABOLISM | 198 | 112 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | 31 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME PI METABOLISM | 48 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | 9 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K AKT ACTIVATION | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME TCR SIGNALING | 54 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM TCR SIGNALING | 37 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME GAB1 SIGNALOSOME | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME PI 3K CASCADE | 56 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | 100 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME PIP3 ACTIVATES AKT SIGNALING | 29 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 UP | 120 | 73 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK DN | 137 | 97 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| HWANG PROSTATE CANCER MARKERS | 28 | 19 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 XPCS DN | 88 | 71 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER DN | 54 | 37 | All SZGR 2.0 genes in this pathway |
| SEIDEN ONCOGENESIS BY MET | 88 | 53 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| HOWLIN PUBERTAL MAMMARY GLAND | 69 | 40 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| COURTOIS SENESCENCE TRIGGERS | 6 | 5 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 7 | 27 | 22 | All SZGR 2.0 genes in this pathway |
| TCGA GLIOBLASTOMA COPY NUMBER DN | 31 | 21 | All SZGR 2.0 genes in this pathway |
| LANDEMAINE LUNG METASTASIS | 21 | 12 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS UP | 81 | 54 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| JAZAERI BREAST CANCER BRCA1 VS BRCA2 DN | 43 | 31 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| DELASERNA MYOD TARGETS DN | 57 | 42 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| KEEN RESPONSE TO ROSIGLITAZONE DN | 106 | 68 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| LIN MELANOMA COPY NUMBER DN | 41 | 34 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN | 154 | 101 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS DN | 120 | 81 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA | 51 | 35 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| EHLERS ANEUPLOIDY UP | 41 | 29 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| PALOMERO GSI SENSITIVITY UP | 7 | 6 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| TCGA GLIOBLASTOMA MUTATED | 8 | 8 | All SZGR 2.0 genes in this pathway |
| KYNG NORMAL AGING DN | 30 | 13 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM DN | 29 | 14 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 3 | 15 | 7 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-130/301 | 2253 | 2259 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-132/212 | 1246 | 1252 | 1A | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC | ||||
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC | ||||
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-141/200a | 1467 | 1473 | m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG | ||||
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-144 | 2917 | 2923 | m8 | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-148/152 | 2254 | 2260 | m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-17-5p/20/93.mr/106/519.d | 272 | 278 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-181 | 2310 | 2316 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-186 | 2743 | 2749 | m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU | ||||
| miR-188 | 762 | 769 | 1A,m8 | hsa-miR-188 | CAUCCCUUGCAUGGUGGAGGGU |
| hsa-miR-188 | CAUCCCUUGCAUGGUGGAGGGU | ||||
| miR-19 | 1221 | 1228 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-193 | 932 | 938 | m8 | hsa-miR-193a | AACUGGCCUACAAAGUCCCAG |
| hsa-miR-193b | AACUGGCCCUCAAAGUCCCGCUUU | ||||
| miR-200bc/429 | 2516 | 2522 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-216 | 1105 | 1111 | m8 | hsa-miR-216 | UAAUCUCAGCUGGCAACUGUG |
| miR-217 | 1330 | 1336 | 1A | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-22 | 689 | 696 | 1A,m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-23 | 1608 | 1615 | 1A,m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC | ||||
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-25/32/92/363/367 | 2859 | 2866 | 1A,m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-26 | 2619 | 2626 | 1A,m8 | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC | ||||
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC | ||||
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-29 | 1741 | 1747 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU | ||||
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-30-3p | 2475 | 2481 | m8 | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC | ||||
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-320 | 2804 | 2811 | 1A,m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-323 | 2924 | 2930 | m8 | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| hsa-miR-323brain | GCACAUUACACGGUCGACCUCU | ||||
| hsa-miR-323brain | GCACAUUACACGGUCGACCUCU | ||||
| miR-328 | 2216 | 2222 | m8 | hsa-miR-328brain | CUGGCCCUCUCUGCCCUUCCGU |
| miR-365 | 2378 | 2384 | 1A | hsa-miR-365 | UAAUGCCCCUAAAAAUCCUUAU |
| miR-369-3p | 2389 | 2395 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU | ||||
| miR-374 | 2389 | 2395 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG | ||||
| miR-410 | 2391 | 2397 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| hsa-miR-410 | AAUAUAACACAGAUGGCCUGU | ||||
| miR-485-5p | 3096 | 3102 | m8 | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
| miR-486 | 3186 | 3193 | 1A,m8 | hsa-miR-486 | UCCUGUACUGAGCUGCCCCGAG |
| hsa-miR-486 | UCCUGUACUGAGCUGCCCCGAG | ||||
| miR-494 | 2313 | 2320 | 1A,m8 | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
| hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU | ||||
| miR-495 | 3232 | 3239 | 1A,m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU | ||||
| hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU | ||||
| miR-542-3p | 353 | 359 | m8 | hsa-miR-542-3p | UGUGACAGAUUGAUAACUGAAA |
| miR-543 | 2311 | 2317 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.