Gene Page: PTPN13
Summary ?
| GeneID | 5783 |
| Symbol | PTPN13 |
| Synonyms | FAP-1|PNP1|PTP-BAS|PTP-BL|PTP1E|PTPL1|PTPLE|hPTP1E |
| Description | protein tyrosine phosphatase, non-receptor type 13 |
| Reference | MIM:600267|HGNC:HGNC:9646|Ensembl:ENSG00000163629|HPRD:02602|Vega:OTTHUMG00000160968 |
| Gene type | protein-coding |
| Map location | 4q21.3 |
| Pascal p-value | 0.579 |
| Sherlock p-value | 8.849E-6 |
| Fetal beta | -0.147 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cerebellum Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg19429466 | 4 | 87515395 | PTPN13 | 4.24E-5 | -0.286 | 0.021 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2976809 | chr3 | 125451738 | PTPN13 | 5783 | 0.19 | trans | ||
| rs4743989 | chr9 | 97859191 | PTPN13 | 5783 | 0.18 | trans |
Section II. Transcriptome annotation
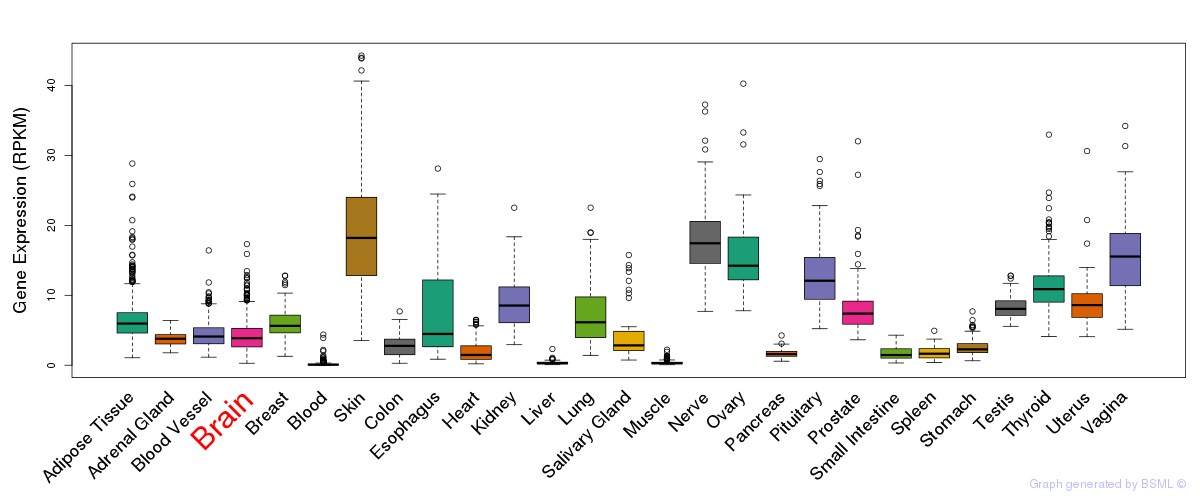
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APC | BTPS2 | DP2 | DP2.5 | DP3 | GS | adenomatous polyposis coli | Affinity Capture-Western Two-hybrid | BioGRID | 10951583 |
| ARHGAP29 | PARG1 | RP11-255E17.1 | Rho GTPase activating protein 29 | - | HPRD,BioGRID | 9305890 |
| BRD7 | BP75 | CELTIX1 | NAG4 | bromodomain containing 7 | - | HPRD | 10526152 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Reconstituted Complex | BioGRID | 10951583 |
| EFNB1 | CFND | CFNS | EFL3 | EPLG2 | Elk-L | LERK2 | MGC8782 | ephrin-B1 | - | HPRD,BioGRID | 9920925 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | - | HPRD | 7536343 |9079683 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | - | HPRD,BioGRID | 9261095 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | - | HPRD,BioGRID | 9882613 |
| NGFRAP1 | BEX3 | Bex | DXS6984E | HGR74 | NADE | nerve growth factor receptor (TNFRSF16) associated protein 1 | - | HPRD | 10544233 |
| PKN2 | MGC150606 | MGC71074 | PAK2 | PRK2 | PRKCL2 | PRO2042 | Pak-2 | protein kinase N2 | - | HPRD,BioGRID | 11356191 |
| RAPGEF6 | DKFZp667N084 | DKFZp686I15116 | KIA001LB | PDZ-GEF2 | PDZGEF2 | RA-GEF-2 | Rap guanine nucleotide exchange factor (GEF) 6 | - | HPRD | 12095257 |
| TRIP6 | MGC10556 | MGC10558 | MGC29959 | MGC3837 | MGC4423 | OIP1 | ZRP-1 | thyroid hormone receptor interactor 6 | - | HPRD,BioGRID | 10826496 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD | 9305890 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA FAS PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID FCER1 PATHWAY | 62 | 43 | All SZGR 2.0 genes in this pathway |
| PID EPHRINB REV PATHWAY | 30 | 25 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN | 182 | 111 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION UP | 119 | 66 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 6HR UP | 85 | 54 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 3D UP | 182 | 110 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| KORKOLA YOLK SAC TUMOR | 62 | 33 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS UP | 81 | 54 | All SZGR 2.0 genes in this pathway |
| NUMATA CSF3 SIGNALING VIA STAT3 | 22 | 17 | All SZGR 2.0 genes in this pathway |
| NGUYEN NOTCH1 TARGETS DN | 86 | 67 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR DN | 101 | 70 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN | 91 | 64 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P3 | 160 | 103 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS DN | 207 | 139 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 4 | 110 | 66 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA NS UP | 162 | 103 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |