Gene Page: PRODH2
Summary ?
| GeneID | 58510 |
| Symbol | PRODH2 |
| Synonyms | HSPOX1|HYPDH |
| Description | proline dehydrogenase (oxidase) 2 |
| Reference | MIM:616377|HGNC:HGNC:17325|Ensembl:ENSG00000250799|HPRD:17911|Vega:OTTHUMG00000180688 |
| Gene type | protein-coding |
| Map location | 19q13.1 |
| Pascal p-value | 0.827 |
| Fetal beta | -0.422 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
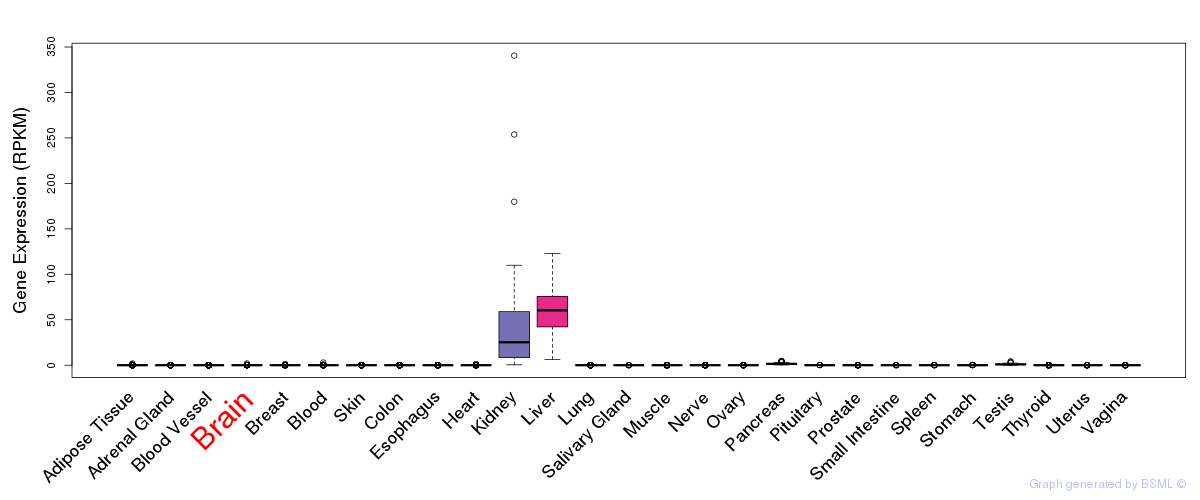
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NDUFA2 | 0.92 | 0.80 |
| UQCRQ | 0.91 | 0.80 |
| LSMD1 | 0.91 | 0.77 |
| MYEOV2 | 0.91 | 0.77 |
| ATP5J2 | 0.90 | 0.81 |
| COX7A2 | 0.90 | 0.82 |
| TIMM8B | 0.90 | 0.84 |
| MRPL52 | 0.90 | 0.76 |
| NDUFB7 | 0.90 | 0.79 |
| MRPL54 | 0.90 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MYH9 | -0.49 | -0.45 |
| KIF16B | -0.46 | -0.46 |
| SLC6A6 | -0.45 | -0.47 |
| BAT2D1 | -0.43 | -0.41 |
| LRP1 | -0.43 | -0.37 |
| MTR | -0.42 | -0.42 |
| PPFIA1 | -0.42 | -0.33 |
| MDN1 | -0.42 | -0.38 |
| C14orf43 | -0.42 | -0.40 |
| MAGI2 | -0.42 | -0.37 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| GO:0004657 | proline dehydrogenase activity | IEA | - | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006537 | glutamate biosynthetic process | IEA | glutamate (GO term level: 9) | - |
| GO:0007254 | JNK cascade | IEA | - | |
| GO:0006562 | proline catabolic process | IEA | - | |
| GO:0055114 | oxidation reduction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ARGININE AND PROLINE METABOLISM | 54 | 39 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G123 DN | 51 | 30 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS DN | 149 | 85 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |