Gene Page: BCR
Summary ?
| GeneID | 613 |
| Symbol | BCR |
| Synonyms | ALL|BCR1|CML|D22S11|D22S662|PHL |
| Description | breakpoint cluster region |
| Reference | MIM:151410|HGNC:HGNC:1014|Ensembl:ENSG00000186716|HPRD:01044|Vega:OTTHUMG00000150655 |
| Gene type | protein-coding |
| Map location | 22q11.23 |
| Pascal p-value | 0.773 |
| Sherlock p-value | 0.524 |
| Fetal beta | -0.386 |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1296 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs131684 | 22 | 23646257 | BCR | ENSG00000186716.15 | 1.785E-8 | 0 | 124366 | gtex_brain_ba24 |
| rs140507 | 22 | 23646512 | BCR | ENSG00000186716.15 | 1.785E-8 | 0 | 124621 | gtex_brain_ba24 |
| rs131686 | 22 | 23646953 | BCR | ENSG00000186716.15 | 1.785E-8 | 0 | 125062 | gtex_brain_ba24 |
| rs131689 | 22 | 23647845 | BCR | ENSG00000186716.15 | 1.788E-7 | 0 | 125954 | gtex_brain_ba24 |
| rs131690 | 22 | 23648009 | BCR | ENSG00000186716.15 | 1.786E-8 | 0 | 126118 | gtex_brain_ba24 |
| rs131691 | 22 | 23648625 | BCR | ENSG00000186716.15 | 1.51E-8 | 0 | 126734 | gtex_brain_ba24 |
| rs140508 | 22 | 23648913 | BCR | ENSG00000186716.15 | 1.785E-7 | 0 | 127022 | gtex_brain_ba24 |
| rs131693 | 22 | 23649242 | BCR | ENSG00000186716.15 | 1.796E-8 | 0 | 127351 | gtex_brain_ba24 |
| rs149904420 | 22 | 23649868 | BCR | ENSG00000186716.15 | 3.245E-8 | 0 | 127977 | gtex_brain_ba24 |
| rs28405226 | 22 | 23649994 | BCR | ENSG00000186716.15 | 1.225E-8 | 0 | 128103 | gtex_brain_ba24 |
| rs131703 | 22 | 23652201 | BCR | ENSG00000186716.15 | 1.294E-7 | 0 | 130310 | gtex_brain_ba24 |
| rs140509 | 22 | 23652212 | BCR | ENSG00000186716.15 | 1.299E-7 | 0 | 130321 | gtex_brain_ba24 |
| rs180797 | 22 | 23652956 | BCR | ENSG00000186716.15 | 1.944E-7 | 0 | 131065 | gtex_brain_ba24 |
| rs180801 | 22 | 23653880 | BCR | ENSG00000186716.15 | 1.246E-7 | 0 | 131989 | gtex_brain_ba24 |
| rs180805 | 22 | 23654993 | BCR | ENSG00000186716.15 | 5.211E-8 | 0 | 133102 | gtex_brain_ba24 |
| rs180806 | 22 | 23655219 | BCR | ENSG00000186716.15 | 1.317E-7 | 0 | 133328 | gtex_brain_ba24 |
| rs180810 | 22 | 23657477 | BCR | ENSG00000186716.15 | 1.273E-7 | 0 | 135586 | gtex_brain_ba24 |
| rs180811 | 22 | 23657495 | BCR | ENSG00000186716.15 | 1.371E-6 | 0 | 135604 | gtex_brain_ba24 |
| rs180812 | 22 | 23657735 | BCR | ENSG00000186716.15 | 1.457E-7 | 0 | 135844 | gtex_brain_ba24 |
| rs392389 | 22 | 23657874 | BCR | ENSG00000186716.15 | 1.507E-7 | 0 | 135983 | gtex_brain_ba24 |
| rs440531 | 22 | 23657875 | BCR | ENSG00000186716.15 | 1.507E-7 | 0 | 135984 | gtex_brain_ba24 |
| rs113629387 | 22 | 23657939 | BCR | ENSG00000186716.15 | 5.657E-7 | 0 | 136048 | gtex_brain_ba24 |
| rs28379041 | 22 | 23657966 | BCR | ENSG00000186716.15 | 1.469E-7 | 0 | 136075 | gtex_brain_ba24 |
| rs180816 | 22 | 23657980 | BCR | ENSG00000186716.15 | 1.29E-7 | 0 | 136089 | gtex_brain_ba24 |
| rs180817 | 22 | 23658007 | BCR | ENSG00000186716.15 | 1.182E-7 | 0 | 136116 | gtex_brain_ba24 |
| rs201326729 | 22 | 23662657 | BCR | ENSG00000186716.15 | 5.284E-7 | 0 | 140766 | gtex_brain_ba24 |
| rs12163260 | 22 | 23662671 | BCR | ENSG00000186716.15 | 5.636E-8 | 0 | 140780 | gtex_brain_ba24 |
| rs4632463 | 22 | 23662897 | BCR | ENSG00000186716.15 | 5.757E-7 | 0 | 141006 | gtex_brain_ba24 |
| rs4822384 | 22 | 23663848 | BCR | ENSG00000186716.15 | 2.012E-7 | 0 | 141957 | gtex_brain_ba24 |
| rs131684 | 22 | 23646257 | BCR | ENSG00000186716.15 | 6.52E-8 | 0 | 124366 | gtex_brain_putamen_basal |
| rs140507 | 22 | 23646512 | BCR | ENSG00000186716.15 | 5.125E-8 | 0 | 124621 | gtex_brain_putamen_basal |
| rs131686 | 22 | 23646953 | BCR | ENSG00000186716.15 | 3.399E-8 | 0 | 125062 | gtex_brain_putamen_basal |
| rs131689 | 22 | 23647845 | BCR | ENSG00000186716.15 | 6.637E-8 | 0 | 125954 | gtex_brain_putamen_basal |
| rs131690 | 22 | 23648009 | BCR | ENSG00000186716.15 | 1.56E-8 | 0 | 126118 | gtex_brain_putamen_basal |
| rs131691 | 22 | 23648625 | BCR | ENSG00000186716.15 | 1.37E-8 | 0 | 126734 | gtex_brain_putamen_basal |
| rs140508 | 22 | 23648913 | BCR | ENSG00000186716.15 | 6.63E-8 | 0 | 127022 | gtex_brain_putamen_basal |
| rs131693 | 22 | 23649242 | BCR | ENSG00000186716.15 | 1.505E-8 | 0 | 127351 | gtex_brain_putamen_basal |
| rs149904420 | 22 | 23649868 | BCR | ENSG00000186716.15 | 4.142E-8 | 0 | 127977 | gtex_brain_putamen_basal |
| rs28405226 | 22 | 23649994 | BCR | ENSG00000186716.15 | 1.377E-8 | 0 | 128103 | gtex_brain_putamen_basal |
| rs131703 | 22 | 23652201 | BCR | ENSG00000186716.15 | 8.154E-13 | 0 | 130310 | gtex_brain_putamen_basal |
| rs140509 | 22 | 23652212 | BCR | ENSG00000186716.15 | 8.101E-13 | 0 | 130321 | gtex_brain_putamen_basal |
| rs180797 | 22 | 23652956 | BCR | ENSG00000186716.15 | 1.237E-12 | 0 | 131065 | gtex_brain_putamen_basal |
| rs180801 | 22 | 23653880 | BCR | ENSG00000186716.15 | 1.153E-12 | 0 | 131989 | gtex_brain_putamen_basal |
| rs56024092 | 22 | 23654807 | BCR | ENSG00000186716.15 | 1.382E-6 | 0 | 132916 | gtex_brain_putamen_basal |
| rs180805 | 22 | 23654993 | BCR | ENSG00000186716.15 | 1.233E-12 | 0 | 133102 | gtex_brain_putamen_basal |
| rs11558697 | 22 | 23655084 | BCR | ENSG00000186716.15 | 1.155E-6 | 0 | 133193 | gtex_brain_putamen_basal |
| rs180806 | 22 | 23655219 | BCR | ENSG00000186716.15 | 3.123E-12 | 0 | 133328 | gtex_brain_putamen_basal |
| rs66569843 | 22 | 23656148 | BCR | ENSG00000186716.15 | 1.109E-6 | 0 | 134257 | gtex_brain_putamen_basal |
| rs180809 | 22 | 23657326 | BCR | ENSG00000186716.15 | 5.684E-8 | 0 | 135435 | gtex_brain_putamen_basal |
| rs180810 | 22 | 23657477 | BCR | ENSG00000186716.15 | 2.953E-9 | 0 | 135586 | gtex_brain_putamen_basal |
| rs180811 | 22 | 23657495 | BCR | ENSG00000186716.15 | 8.444E-11 | 0 | 135604 | gtex_brain_putamen_basal |
| rs180812 | 22 | 23657735 | BCR | ENSG00000186716.15 | 3.142E-12 | 0 | 135844 | gtex_brain_putamen_basal |
| rs392389 | 22 | 23657874 | BCR | ENSG00000186716.15 | 3.057E-12 | 0 | 135983 | gtex_brain_putamen_basal |
| rs440531 | 22 | 23657875 | BCR | ENSG00000186716.15 | 3.057E-12 | 0 | 135984 | gtex_brain_putamen_basal |
| rs113629387 | 22 | 23657939 | BCR | ENSG00000186716.15 | 3.868E-12 | 0 | 136048 | gtex_brain_putamen_basal |
| rs28379041 | 22 | 23657966 | BCR | ENSG00000186716.15 | 1.227E-8 | 0 | 136075 | gtex_brain_putamen_basal |
| rs180816 | 22 | 23657980 | BCR | ENSG00000186716.15 | 6.116E-9 | 0 | 136089 | gtex_brain_putamen_basal |
| rs180817 | 22 | 23658007 | BCR | ENSG00000186716.15 | 1.91E-11 | 0 | 136116 | gtex_brain_putamen_basal |
| rs2330350 | 22 | 23660942 | BCR | ENSG00000186716.15 | 1.58E-8 | 0 | 139051 | gtex_brain_putamen_basal |
| rs13057338 | 22 | 23660980 | BCR | ENSG00000186716.15 | 2.706E-10 | 0 | 139089 | gtex_brain_putamen_basal |
| rs9612293 | 22 | 23661273 | BCR | ENSG00000186716.15 | 1.232E-9 | 0 | 139382 | gtex_brain_putamen_basal |
| rs201326729 | 22 | 23662657 | BCR | ENSG00000186716.15 | 1.815E-9 | 0 | 140766 | gtex_brain_putamen_basal |
| rs12163260 | 22 | 23662671 | BCR | ENSG00000186716.15 | 7.83E-10 | 0 | 140780 | gtex_brain_putamen_basal |
| rs4632463 | 22 | 23662897 | BCR | ENSG00000186716.15 | 6.085E-10 | 0 | 141006 | gtex_brain_putamen_basal |
| rs8140083 | 22 | 23663722 | BCR | ENSG00000186716.15 | 1.471E-10 | 0 | 141831 | gtex_brain_putamen_basal |
| rs13053509 | 22 | 23663772 | BCR | ENSG00000186716.15 | 9.671E-7 | 0 | 141881 | gtex_brain_putamen_basal |
| rs4822384 | 22 | 23663848 | BCR | ENSG00000186716.15 | 9.318E-10 | 0 | 141957 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
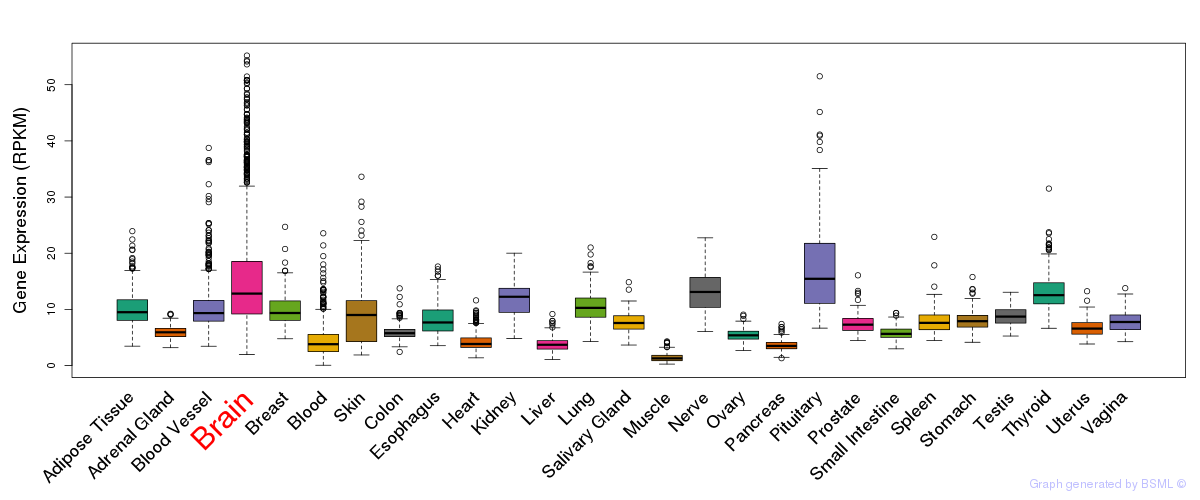
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005089 | Rho guanyl-nucleotide exchange factor activity | IEA | - | |
| GO:0005085 | guanyl-nucleotide exchange factor activity | IEA | - | |
| GO:0005096 | GTPase activator activity | NAS | - | |
| GO:0005096 | GTPase activator activity | TAS | 1903516 | |
| GO:0004674 | protein serine/threonine kinase activity | NAS | - | |
| GO:0004674 | protein serine/threonine kinase activity | TAS | 1657398 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0016301 | kinase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | IEA | Brain (GO term level: 7) | - |
| GO:0006468 | protein amino acid phosphorylation | TAS | 1657398 | |
| GO:0007242 | intracellular signaling cascade | IEA | - | |
| GO:0007242 | intracellular signaling cascade | NAS | - | |
| GO:0035023 | regulation of Rho protein signal transduction | IEA | - | |
| GO:0051726 | regulation of cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005622 | intracellular | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | BCR interacts with c-Abl. This interaction was modeled on a demonstrated interaction between BCR from an unspecified source and human c-Abl. | BIND | 1383690 |
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD | 11780146 |
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | Affinity Capture-Western Reconstituted Complex | BioGRID | 1712671 |8112292 |12543778 |
| ABL2 | ABLL | ARG | FLJ22224 | FLJ31718 | FLJ41441 | v-abl Abelson murine leukemia viral oncogene homolog 2 (arg, Abelson-related gene) | BCR interacts with ARG. This interaction was modeled on a demonstrated interaction between BCR from an unspecified source and human ARG. | BIND | 1383690 |
| BCR | ALL | BCR-ABL1 | BCR1 | CML | D22S11 | D22S662 | FLJ16453 | PHL | breakpoint cluster region | - | HPRD | 11780146 |
| BCR | ALL | BCR-ABL1 | BCR1 | CML | D22S11 | D22S662 | FLJ16453 | PHL | breakpoint cluster region | p210 Bcr-Abl phosphorylates p160 Bcr. | BIND | 8112292 |
| BLK | MGC10442 | B lymphoid tyrosine kinase | - | HPRD | 8756631 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | Co-purification | BioGRID | 8641358 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | Affinity Capture-Western Far Western Two-hybrid | BioGRID | 8978305 |9747873 |10194128 |15143164 |
| ERCC3 | BTF2 | GTF2H | RAD25 | TFIIH | XPB | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | - | HPRD,BioGRID | 9874796 |
| FES | FPS | feline sarcoma oncogene | - | HPRD,BioGRID | 7529874 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | Affinity Capture-Western | BioGRID | 15143164 |
| GRB10 | GRB-IR | Grb-10 | IRBP | KIAA0207 | MEG1 | RSS | growth factor receptor-bound protein 10 | - | HPRD,BioGRID | 9747873 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Grb2 interacts with Bcr-Abl. | BIND | 9872323 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 8112292 |9178913 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Tyrosine phosphorylated Bcr-Abl interacts with Grb2. | BIND | 8112292 |
| HCK | JTK9 | hemopoietic cell kinase | Affinity Capture-Western | BioGRID | 10849448 |12592324 |
| HOXA9 | ABD-B | HOX1 | HOX1.7 | HOX1G | MGC1934 | homeobox A9 | Phenotypic Enhancement | BioGRID | 12393433 |
| IGSF21 | FLJ41177 | MGC15730 | immunoglobin superfamily, member 21 | Two-hybrid | BioGRID | 16169070 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | - | HPRD,BioGRID | 12023981 |12370803 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | Affinity Capture-Western | BioGRID | 8757502 |
| MAP4K5 | GCKR | KHS | KHS1 | MAPKKKK5 | mitogen-activated protein kinase kinase kinase kinase 5 | - | HPRD | 9949177 |
| MLLT4 | AF-6 | AF6 | AFADIN | FLJ34371 | RP3-431P23.3 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 | - | HPRD,BioGRID | 12808105 |
| NCK2 | GRB4 | NCKbeta | NCK adaptor protein 2 | - | HPRD | 10887126 |
| PIK3CG | PI3CG | PI3K | PI3Kgamma | PIK3 | phosphoinositide-3-kinase, catalytic, gamma polypeptide | Affinity Capture-Western Co-purification Two-hybrid | BioGRID | 7606002 |8641358 |9747873 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | BCR interacts with PLCgamma1. This interaction was modeled on a demonstrated interaction between BCR from an unspecified source and bovine PLCgamma1. | BIND | 1383690 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | Phosphorylated p210bcr-abl interacts with phosphorylated Syp. This interaction was modeled on a demonstrated interaction between human p210bcr-abl and mouse Syp. | BIND | 8195176 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | - | HPRD | 12115002 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | - | HPRD,BioGRID | 9211920 |
| PXN | FLJ16691 | paxillin | Affinity Capture-Western Co-purification | BioGRID | 7534286 |8641358 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | BCR interacts with GAP. This interaction was modeled on a demonstrated interaction between BCR from an unspecified source and human GAP. | BIND | 1383690 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Affinity Capture-Western | BioGRID | 9071815 |
| SH2B1 | DKFZp547G1110 | FLJ30542 | KIAA1299 | SH2-B | SH2B | SH2B adaptor protein 1 | Affinity Capture-Western | BioGRID | 12370803 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 9209414 |
| SOS1 | GF1 | GGF1 | GINGF | HGF | NS4 | son of sevenless homolog 1 (Drosophila) | Affinity Capture-Western | BioGRID | 8112292 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | BCR interacts with Src. This interaction was modeled on a demonstrated interaction between BCR from an unspecified source and Src from Rous Sarcoma virus. | BIND | 1383690 |
| TLN1 | ILWEQ | KIAA1027 | TLN | talin 1 | Co-purification | BioGRID | 8641358 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Two-hybrid | BioGRID | 16169070 |
| UNC119 | HRG4 | unc-119 homolog (C. elegans) | Two-hybrid | BioGRID | 16169070 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD | 11790798 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD | 7939633 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GLEEVEC PATHWAY | 23 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTEGRIN PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| SA B CELL RECEPTOR COMPLEXES | 24 | 20 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN B LYMPHOCYTES | 36 | 31 | All SZGR 2.0 genes in this pathway |
| SIG BCR SIGNALING PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| ST B CELL ANTIGEN RECEPTOR | 40 | 32 | All SZGR 2.0 genes in this pathway |
| PID RHOA REG PATHWAY | 46 | 30 | All SZGR 2.0 genes in this pathway |
| PID RAC1 REG PATHWAY | 38 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY RHO GTPASES | 113 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR1 MUTANTS | 30 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | 19 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR MUTANTS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA DN | 116 | 79 | All SZGR 2.0 genes in this pathway |
| LIU CMYB TARGETS UP | 165 | 106 | All SZGR 2.0 genes in this pathway |
| LIU VMYB TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| BARRIER CANCER RELAPSE NORMAL SAMPLE DN | 31 | 19 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 22 | 13 | 9 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE AUGMENTED BY MYC | 108 | 74 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH3 21 UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| NUTT GBM VS AO GLIOMA DN | 45 | 22 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| ZHAN VARIABLE EARLY DIFFERENTIATION GENES UP | 15 | 9 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| LINSLEY MIR16 TARGETS | 206 | 127 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN ASPARAGINASE RESISTANCE B ALL UP | 26 | 15 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN ASPARAGINASE RESISTANCE ALL UP | 22 | 14 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-145 | 2008 | 2015 | 1A,m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-15/16/195/424/497 | 419 | 425 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-181 | 1059 | 1065 | 1A | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-26 | 983 | 990 | 1A,m8 | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-96 | 2034 | 2040 | m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.