Gene Page: PDLIM2
Summary ?
| GeneID | 64236 |
| Symbol | PDLIM2 |
| Synonyms | MYSTIQUE|SLIM |
| Description | PDZ and LIM domain 2 |
| Reference | MIM:609722|HGNC:HGNC:13992|Ensembl:ENSG00000120913|HPRD:17831|Vega:OTTHUMG00000164270 |
| Gene type | protein-coding |
| Map location | 8p21.3 |
| Pascal p-value | 0.133 |
| Sherlock p-value | 0.004 |
| Fetal beta | -0.856 |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.03086 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.00057 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs881771 | chr8 | 22431587 | PDLIM2 | 64236 | 0.01 | cis | ||
| rs3064 | chr8 | 22451687 | PDLIM2 | 64236 | 3.233E-12 | cis | ||
| rs3735893 | chr8 | 22452703 | PDLIM2 | 64236 | 3.952E-12 | cis | ||
| rs2291232 | chr8 | 22464063 | PDLIM2 | 64236 | 7.363E-11 | cis | ||
| rs1871900 | chr8 | 22481448 | PDLIM2 | 64236 | 7.45E-7 | cis | ||
| rs10099266 | chr8 | 22484267 | PDLIM2 | 64236 | 5.98E-7 | cis | ||
| rs6558172 | chr8 | 22492051 | PDLIM2 | 64236 | 0.2 | cis | ||
| rs6558173 | chr8 | 22492102 | PDLIM2 | 64236 | 4.967E-10 | cis | ||
| rs11776549 | chr8 | 22493123 | PDLIM2 | 64236 | 1.71E-9 | cis | ||
| rs7005025 | chr8 | 22519814 | PDLIM2 | 64236 | 3.226E-9 | cis | ||
| rs10104039 | chr8 | 22538383 | PDLIM2 | 64236 | 2.537E-7 | cis | ||
| rs1040053 | chr8 | 22699218 | PDLIM2 | 64236 | 0.17 | cis | ||
| rs16829545 | chr2 | 151977407 | PDLIM2 | 64236 | 0 | trans | ||
| rs3064 | chr8 | 22451687 | PDLIM2 | 64236 | 8.123E-10 | trans | ||
| rs3735893 | chr8 | 22452703 | PDLIM2 | 64236 | 9.952E-10 | trans | ||
| rs2291232 | chr8 | 22464063 | PDLIM2 | 64236 | 1.897E-8 | trans | ||
| rs1871900 | chr8 | 22481448 | PDLIM2 | 64236 | 1.42E-4 | trans | ||
| rs10099266 | chr8 | 22484267 | PDLIM2 | 64236 | 1.169E-4 | trans | ||
| rs6558173 | chr8 | 22492102 | PDLIM2 | 64236 | 1.209E-7 | trans | ||
| rs11776549 | chr8 | 22493123 | PDLIM2 | 64236 | 4.157E-7 | trans | ||
| rs7005025 | chr8 | 22519814 | PDLIM2 | 64236 | 7.706E-7 | trans | ||
| rs10104039 | chr8 | 22538383 | PDLIM2 | 64236 | 5.284E-5 | trans | ||
| rs16955618 | chr15 | 29937543 | PDLIM2 | 64236 | 0 | trans |
Section II. Transcriptome annotation
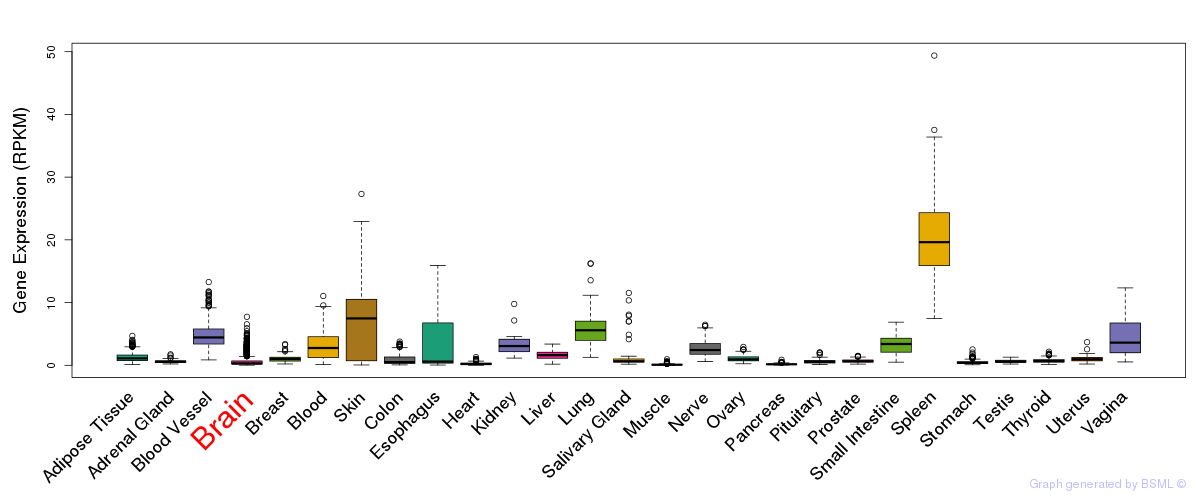
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0009986 | cell surface | IDA | 12493773 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| FURUKAWA DUSP6 TARGETS PCI35 DN | 74 | 40 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| FLOTHO PEDIATRIC ALL THERAPY RESPONSE DN | 29 | 17 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS DN | 23 | 16 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| STEGER ADIPOGENESIS DN | 25 | 15 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-299-3p | 2971 | 2977 | m8 | hsa-miR-299-3p | UAUGUGGGAUGGUAAACCGCUU |
| miR-491 | 2970 | 2976 | 1A | hsa-miR-491brain | AGUGGGGAACCCUUCCAUGAGGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.