Gene Page: DCLRE1C
Summary ?
| GeneID | 64421 |
| Symbol | DCLRE1C |
| Synonyms | A-SCID|DCLREC1C|RS-SCID|SCIDA|SNM1C |
| Description | DNA cross-link repair 1C |
| Reference | MIM:605988|HGNC:HGNC:17642|Ensembl:ENSG00000152457|HPRD:05817|Vega:OTTHUMG00000017716 |
| Gene type | protein-coding |
| Map location | 10p13 |
| Pascal p-value | 0.19 |
| Sherlock p-value | 0.365 |
| Fetal beta | 1.79 |
| DMG | 2 (# studies) |
| eGene | Cerebellar Hemisphere Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| Expression | Meta-analysis of gene expression | P value: 2.463 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09688494 | 10 | 14996222 | DCLRE1C | 2.239E-4 | -0.244 | 0.036 | DMG:Wockner_2014 |
| cg11038604 | 10 | 14996337 | DCLRE1C | 2.06E-8 | -0.012 | 7.09E-6 | DMG:Jaffe_2016 |
| cg04785461 | 10 | 14996236 | DCLRE1C | 3.65E-8 | -0.007 | 1.06E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| snp_a-1878583 | 0 | DCLRE1C | 64421 | 0.09 | trans |
Section II. Transcriptome annotation
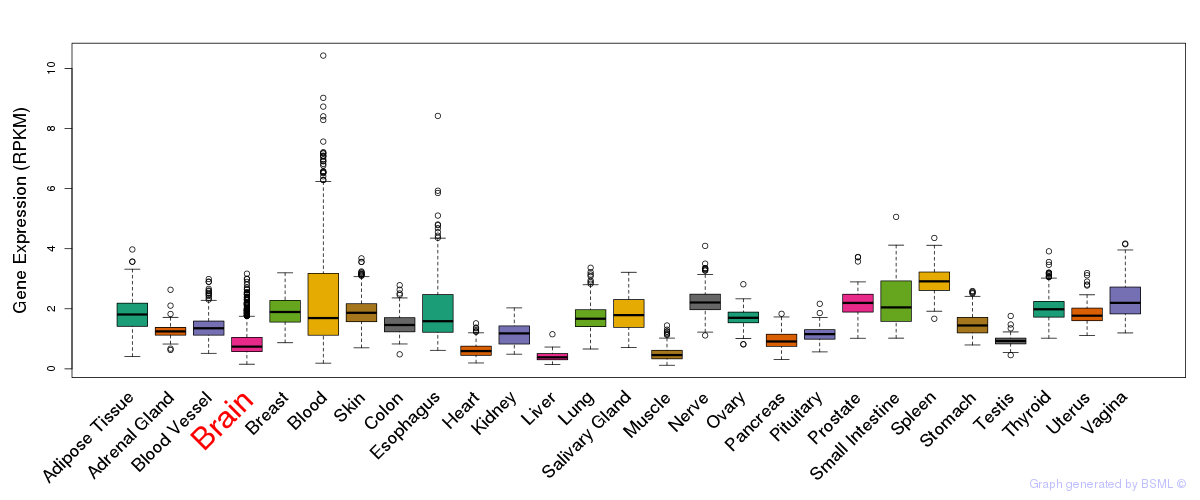
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0004519 | endonuclease activity | IEA | - | |
| GO:0004527 | exonuclease activity | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006310 | DNA recombination | IEA | - | |
| GO:0006281 | DNA repair | IEA | - | |
| GO:0006955 | immune response | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NON HOMOLOGOUS END JOINING | 14 | 8 | All SZGR 2.0 genes in this pathway |
| KEGG PRIMARY IMMUNODEFICIENCY | 35 | 28 | All SZGR 2.0 genes in this pathway |
| PID DNA PK PATHWAY | 16 | 10 | All SZGR 2.0 genes in this pathway |
| PID ATM PATHWAY | 34 | 25 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA MYELOID DN | 38 | 25 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| COLLIS PRKDC SUBSTRATES | 20 | 15 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR UP | 223 | 132 | All SZGR 2.0 genes in this pathway |
| SPIRA SMOKERS LUNG CANCER DN | 22 | 11 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS DN | 187 | 109 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |
| ZWANG DOWN BY 2ND EGF PULSE | 293 | 119 | All SZGR 2.0 genes in this pathway |