Gene Page: REEP1
Summary ?
| GeneID | 65055 |
| Symbol | REEP1 |
| Synonyms | C2orf23|HMN5B|SPG31|Yip2a |
| Description | receptor accessory protein 1 |
| Reference | MIM:609139|HGNC:HGNC:25786|Ensembl:ENSG00000068615|HPRD:07810|Vega:OTTHUMG00000130205 |
| Gene type | protein-coding |
| Map location | 2p11.2 |
| Pascal p-value | 0.305 |
| Sherlock p-value | 0.795 |
| Fetal beta | 1.081 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0004 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21569844 | 2 | 86564582 | REEP1 | 3.37E-8 | -0.016 | 1.01E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | REEP1 | 65055 | 0.17 | trans | ||
| rs71377068 | 2 | 86477840 | REEP1 | ENSG00000068615.12 | 3.588E-6 | 0.02 | 87366 | gtex_brain_putamen_basal |
| rs397873792 | 2 | 86494130 | REEP1 | ENSG00000068615.12 | 3.307E-6 | 0.02 | 71076 | gtex_brain_putamen_basal |
| rs2367233 | 2 | 86541666 | REEP1 | ENSG00000068615.12 | 1.065E-6 | 0.02 | 23540 | gtex_brain_putamen_basal |
| rs6746198 | 2 | 86542113 | REEP1 | ENSG00000068615.12 | 1.314E-6 | 0.02 | 23093 | gtex_brain_putamen_basal |
| rs11691122 | 2 | 86543595 | REEP1 | ENSG00000068615.12 | 2.841E-6 | 0.02 | 21611 | gtex_brain_putamen_basal |
| rs10200619 | 2 | 86548848 | REEP1 | ENSG00000068615.12 | 1.314E-6 | 0.02 | 16358 | gtex_brain_putamen_basal |
| rs7593440 | 2 | 86550657 | REEP1 | ENSG00000068615.12 | 1.314E-6 | 0.02 | 14549 | gtex_brain_putamen_basal |
| rs4392258 | 2 | 86552577 | REEP1 | ENSG00000068615.12 | 1.314E-6 | 0.02 | 12629 | gtex_brain_putamen_basal |
| rs6547674 | 2 | 86552919 | REEP1 | ENSG00000068615.12 | 2.836E-6 | 0.02 | 12287 | gtex_brain_putamen_basal |
| rs11685863 | 2 | 86554484 | REEP1 | ENSG00000068615.12 | 1.314E-6 | 0.02 | 10722 | gtex_brain_putamen_basal |
| rs11127029 | 2 | 86554625 | REEP1 | ENSG00000068615.12 | 2.97E-6 | 0.02 | 10581 | gtex_brain_putamen_basal |
| rs1863051 | 2 | 86559148 | REEP1 | ENSG00000068615.12 | 2.374E-6 | 0.02 | 6058 | gtex_brain_putamen_basal |
| rs1863052 | 2 | 86559358 | REEP1 | ENSG00000068615.12 | 1.313E-6 | 0.02 | 5848 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
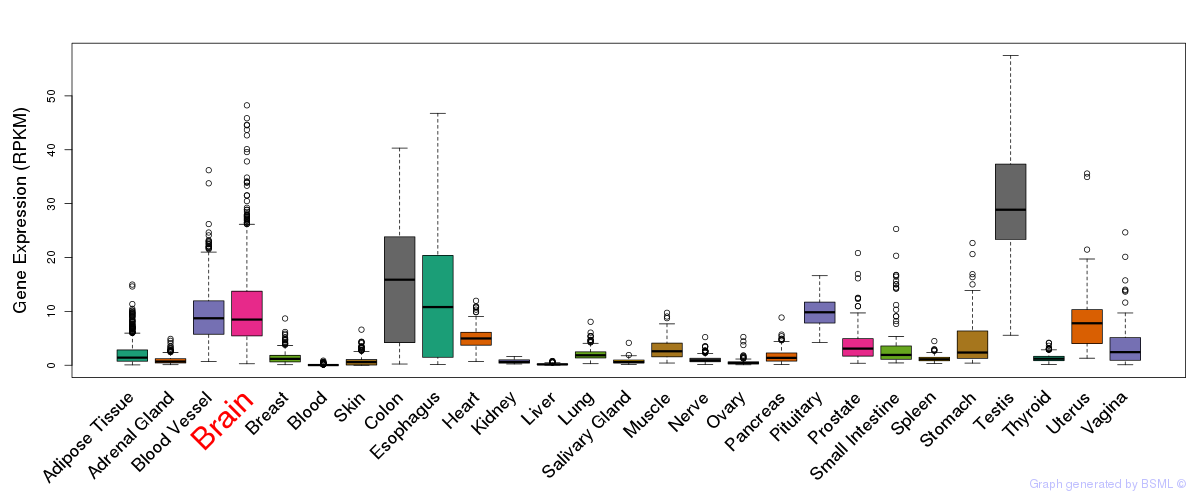
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0031849 | olfactory receptor binding | IMP | 15550249 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0051205 | protein insertion into membrane | IDA | 15550249 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IDA | 15550249 | |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0031966 | mitochondrial membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| WATANABE COLON CANCER MSI VS MSS DN | 81 | 42 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS DN | 68 | 49 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA DN | 284 | 156 | All SZGR 2.0 genes in this pathway |
| SCIBETTA KDM5B TARGETS DN | 81 | 55 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP | 207 | 128 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| KANG GIST WITH PDGFRA UP | 50 | 27 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER C | 113 | 47 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 1 UP | 118 | 66 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS UP | 108 | 75 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD UP | 131 | 87 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN | 245 | 144 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP | 253 | 147 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE | 246 | 153 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE S | 162 | 86 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA | 140 | 96 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 1 | 46 | 33 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 31 | 37 | 1A | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-124.1 | 106 | 113 | 1A,m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 106 | 112 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-140 | 51 | 57 | m8 | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-182 | 2231 | 2237 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-200bc/429 | 2848 | 2854 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-204/211 | 259 | 266 | 1A,m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-208 | 3083 | 3089 | 1A | hsa-miR-208 | AUAAGACGAGCAAAAAGCUUGU |
| miR-214 | 386 | 392 | 1A | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-23 | 2482 | 2489 | 1A,m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-24 | 1460 | 1466 | m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG | ||||
| miR-377 | 2719 | 2726 | 1A,m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-493-5p | 394 | 400 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-495 | 3049 | 3055 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-499 | 3082 | 3089 | 1A,m8 | hsa-miR-499 | UUAAGACUUGCAGUGAUGUUUAA |
| miR-96 | 2231 | 2237 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.