Gene Page: SSTR2
Summary ?
| GeneID | 6752 |
| Symbol | SSTR2 |
| Synonyms | - |
| Description | somatostatin receptor 2 |
| Reference | MIM:182452|HGNC:HGNC:11331|HPRD:01674| |
| Gene type | protein-coding |
| Map location | 17q24 |
| Pascal p-value | 0.943 |
| Fetal beta | 0.473 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06685177 | 17 | 71161322 | SSTR2 | 1.499E-4 | -0.391 | 0.032 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16977927 | chr17 | 72154232 | SSTR2 | 6752 | 0.01 | cis | ||
| rs17638742 | chr3 | 28326697 | SSTR2 | 6752 | 0.01 | trans | ||
| rs11706542 | chr3 | 28328582 | SSTR2 | 6752 | 0.04 | trans | ||
| rs17638991 | chr3 | 28350968 | SSTR2 | 6752 | 0.03 | trans | ||
| rs11720107 | chr3 | 28472727 | SSTR2 | 6752 | 0.07 | trans | ||
| rs17696459 | chr3 | 28491284 | SSTR2 | 6752 | 0.02 | trans | ||
| rs1598846 | chr3 | 28510452 | SSTR2 | 6752 | 0.02 | trans | ||
| rs17696783 | chr3 | 28537610 | SSTR2 | 6752 | 0.06 | trans | ||
| rs7433570 | 0 | SSTR2 | 6752 | 0.02 | trans | |||
| rs4133027 | chr3 | 28624326 | SSTR2 | 6752 | 0.17 | trans | ||
| rs3809540 | chr15 | 55489411 | SSTR2 | 6752 | 0.18 | trans | ||
| rs7165491 | chr15 | 55500454 | SSTR2 | 6752 | 0.13 | trans |
Section II. Transcriptome annotation
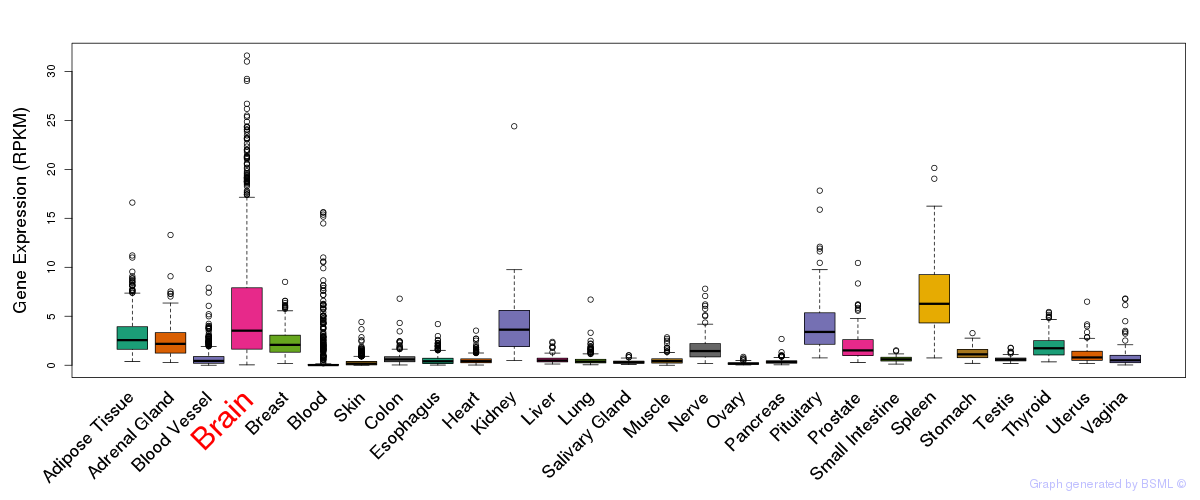
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MYO5A | 0.94 | 0.96 |
| HECW2 | 0.93 | 0.94 |
| LONRF2 | 0.93 | 0.95 |
| SYT16 | 0.93 | 0.93 |
| PCLO | 0.92 | 0.94 |
| FAM126B | 0.91 | 0.94 |
| UBR3 | 0.91 | 0.93 |
| LMTK2 | 0.90 | 0.90 |
| DGKE | 0.90 | 0.89 |
| PLEKHM3 | 0.90 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAB34 | -0.55 | -0.69 |
| EFEMP2 | -0.53 | -0.58 |
| DBI | -0.52 | -0.67 |
| RHOC | -0.51 | -0.65 |
| C19orf36 | -0.49 | -0.58 |
| ACSF2 | -0.49 | -0.62 |
| GATSL3 | -0.48 | -0.57 |
| TLCD1 | -0.48 | -0.58 |
| C1orf61 | -0.48 | -0.73 |
| ACP6 | -0.47 | -0.51 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004994 | somatostatin receptor activity | TAS | 7914078 | |
| GO:0005515 | protein binding | IPI | 10551867 | |
| GO:0030165 | PDZ domain binding | IPI | 10551867 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007218 | neuropeptide signaling pathway | IEA | Neurotransmitter (GO term level: 8) | - |
| GO:0007187 | G-protein signaling, coupled to cyclic nucleotide second messenger | TAS | 7914078 | |
| GO:0007267 | cell-cell signaling | TAS | 1346068 | |
| GO:0007193 | G-protein signaling, adenylate cyclase inhibiting pathway | IEA | - | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0008285 | negative regulation of cell proliferation | TAS | 9892014 | |
| GO:0007586 | digestion | TAS | 1346068 | |
| GO:0007584 | response to nutrient | TAS | 1346068 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005624 | membrane fraction | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0005886 | plasma membrane | TAS | 7914078 | |
| GO:0005887 | integral to plasma membrane | TAS | 7914078 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN | 186 | 107 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1 | 229 | 137 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU UP | 53 | 37 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| NOUZOVA METHYLATED IN APL | 68 | 39 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS UP | 100 | 72 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| OSADA ASCL1 TARGETS UP | 46 | 30 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS HCP WITH H3 UNMETHYLATED | 80 | 50 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES HCP WITH H3 UNMETHYLATED | 63 | 31 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3 UNMETHYLATED | 228 | 119 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS DN | 115 | 73 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| LOPEZ TRANSLATION VIA FN1 SIGNALING | 35 | 21 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| BOSCO TH1 CYTOTOXIC MODULE | 114 | 62 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |