| PID MYC REPRESS PATHWAY
| 63 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA
| 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA
| 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME DESTABILIZATION OF MRNA BY BRF1
| 17 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS
| 84 | 50 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP
| 473 | 314 | All SZGR 2.0 genes in this pathway |
| PRAMOONJAGO SOX4 TARGETS UP
| 52 | 38 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP
| 1382 | 904 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED DN
| 86 | 47 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP
| 306 | 188 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP
| 133 | 78 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP
| 233 | 161 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP
| 456 | 228 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP
| 214 | 155 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN
| 129 | 84 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN
| 209 | 122 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN
| 226 | 132 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS DN
| 120 | 73 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP
| 552 | 347 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP
| 185 | 119 | All SZGR 2.0 genes in this pathway |
| BARRIER CANCER RELAPSE NORMAL SAMPLE DN
| 31 | 19 | All SZGR 2.0 genes in this pathway |
| MANN RESPONSE TO AMIFOSTINE UP
| 20 | 12 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP
| 1037 | 673 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS UP
| 127 | 79 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC UP
| 33 | 20 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC 8HR 5 UP
| 16 | 7 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC 24HR 5 UP
| 34 | 23 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN
| 428 | 306 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN
| 206 | 136 | All SZGR 2.0 genes in this pathway |
| AMIT DELAYED EARLY GENES
| 18 | 15 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS
| 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS
| 290 | 172 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS
| 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH NOS TARGETS
| 179 | 105 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 60 HELA
| 46 | 32 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE UP
| 90 | 62 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE DN
| 121 | 79 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS
| 882 | 572 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP
| 203 | 130 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 DN
| 77 | 48 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS DN
| 32 | 25 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR
| 293 | 193 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP
| 101 | 76 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER HEREDITARY VS SPORADIC
| 50 | 32 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP
| 244 | 165 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN
| 298 | 200 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP
| 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BRACHAT RESPONSE TO METHOTREXATE UP
| 26 | 16 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR DN
| 53 | 40 | All SZGR 2.0 genes in this pathway |
| BRACHAT RESPONSE TO CAMPTOTHECIN UP
| 28 | 18 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP
| 198 | 132 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN
| 128 | 93 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2
| 163 | 113 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL
| 311 | 205 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN
| 96 | 71 | All SZGR 2.0 genes in this pathway |
| SAFFORD T LYMPHOCYTE ANERGY
| 87 | 54 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED
| 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED
| 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS STIMULATED UP
| 29 | 19 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN
| 220 | 147 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY GAMMA IN OLD
| 31 | 19 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY GAMMA IN WS
| 33 | 22 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN
| 195 | 135 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE UP
| 56 | 41 | All SZGR 2.0 genes in this pathway |
| CHENG IMPRINTED BY ESTRADIOL
| 110 | 68 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR UP
| 174 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP
| 973 | 570 | All SZGR 2.0 genes in this pathway |
| PODAR RESPONSE TO ADAPHOSTIN UP
| 147 | 98 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP
| 259 | 185 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP
| 395 | 249 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12
| 317 | 177 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN
| 253 | 192 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS DN
| 31 | 25 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC
| 1103 | 714 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS DN
| 207 | 139 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN
| 505 | 328 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 UP
| 134 | 85 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS DN
| 242 | 146 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP
| 281 | 183 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP
| 745 | 475 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3
| 824 | 528 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4
| 227 | 149 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF
| 222 | 159 | All SZGR 2.0 genes in this pathway |
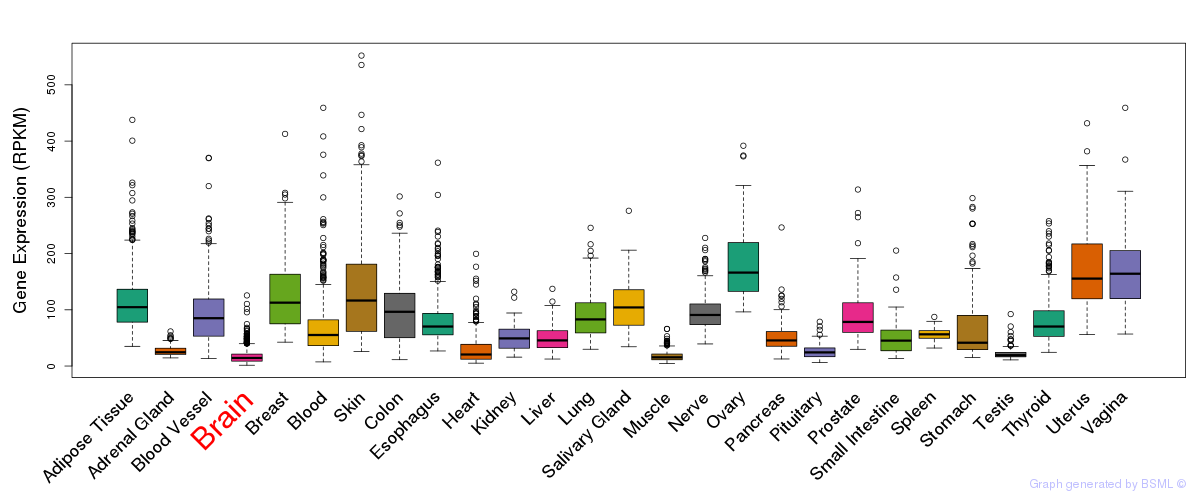
 CV:PheWAS
CV:PheWAS Differentially methylated gene
Differentially methylated gene