Gene Page: TSPO
Summary ?
| GeneID | 706 |
| Symbol | TSPO |
| Synonyms | BPBS|BZRP|DBI|IBP|MBR|PBR|PBS|PKBS|PTBR|mDRC|pk18 |
| Description | translocator protein |
| Reference | MIM:109610|HGNC:HGNC:1158|Ensembl:ENSG00000100300|HPRD:15913|Vega:OTTHUMG00000150573 |
| Gene type | protein-coding |
| Map location | 22q13.31 |
| Pascal p-value | 0.289 |
| Sherlock p-value | 0.156 |
| Fetal beta | 1.107 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| ADT:Sun_2012 | Systematic Investigation of Antipsychotic Drugs and Their Targets | A total of 382 drug-target associations involving 43 antipsychotic drugs and 49 target genes. | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17319860 | chr4 | 160647108 | TSPO | 706 | 0.01 | trans | ||
| rs17153204 | chr10 | 6335730 | TSPO | 706 | 0.03 | trans | ||
| rs3888798 | chr11 | 88027208 | TSPO | 706 | 0.15 | trans | ||
| rs4501739 | chrX | 35960848 | TSPO | 706 | 0.16 | trans | ||
| rs7060495 | chrX | 36100343 | TSPO | 706 | 0.17 | trans |
Section II. Transcriptome annotation
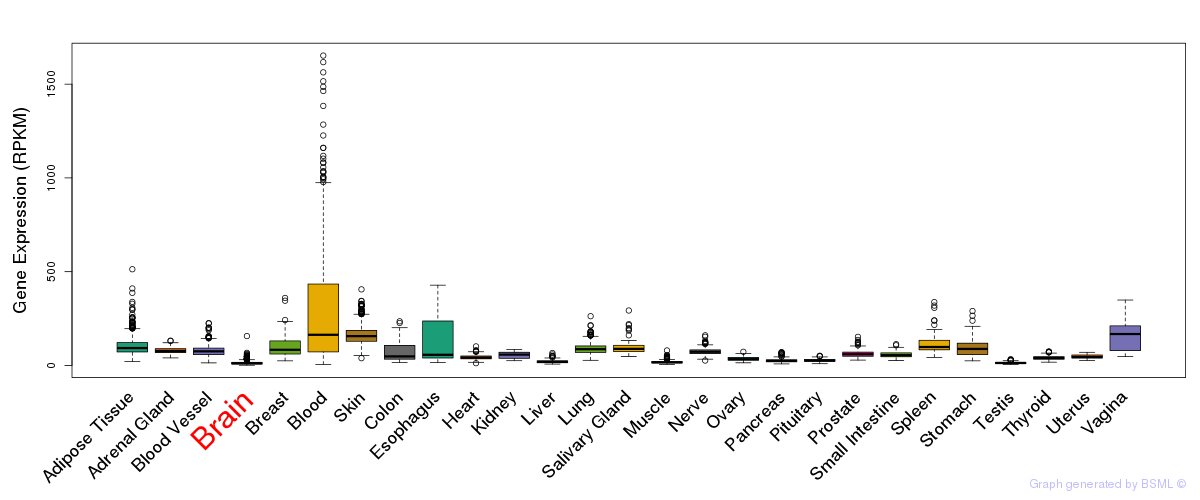
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C1orf85 | 0.69 | 0.65 |
| ILK | 0.67 | 0.56 |
| KIAA0141 | 0.66 | 0.67 |
| TTC38 | 0.64 | 0.63 |
| CPT2 | 0.62 | 0.59 |
| GNG5 | 0.61 | 0.41 |
| C1orf66 | 0.61 | 0.62 |
| TCTN3 | 0.60 | 0.55 |
| TMEM216 | 0.60 | 0.45 |
| DNAJC4 | 0.58 | 0.53 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SEC62 | -0.37 | -0.42 |
| AF347015.21 | -0.30 | -0.25 |
| FAM159B | -0.29 | -0.33 |
| MT-ATP8 | -0.28 | -0.23 |
| AF347015.18 | -0.28 | -0.21 |
| FAM133A | -0.27 | -0.25 |
| AC010300.1 | -0.27 | -0.25 |
| AC098691.2 | -0.27 | -0.22 |
| AC093616.2 | -0.24 | -0.31 |
| AL390816.1 | -0.24 | -0.16 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008503 | benzodiazepine receptor activity | NAS | GABA, Neurotransmitter (GO term level: 6) | 8307574 |
| GO:0008503 | benzodiazepine receptor activity | TAS | GABA, Neurotransmitter (GO term level: 6) | 1326278 |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0003674 | molecular_function | ND | - | |
| GO:0015485 | cholesterol binding | TAS | 16822554 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007165 | signal transduction | NAS | - | |
| GO:0008202 | steroid metabolic process | TAS | 16822554 | |
| GO:0008283 | cell proliferation | TAS | 16822554 | |
| GO:0008150 | biological_process | ND | - | |
| GO:0006820 | anion transport | TAS | 16822554 | |
| GO:0006783 | heme biosynthetic process | TAS | 16822554 | |
| GO:0006626 | protein targeting to mitochondrion | TAS | 1326278 | |
| GO:0006915 | apoptosis | TAS | 16822554 | |
| GO:0032374 | regulation of cholesterol transport | TAS | 16822554 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0005741 | mitochondrial outer membrane | NAS | 8307574 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS CDC25 UP | 58 | 39 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 DN | 153 | 100 | All SZGR 2.0 genes in this pathway |
| BERENJENO ROCK SIGNALING NOT VIA RHOA UP | 29 | 21 | All SZGR 2.0 genes in this pathway |
| CHOI ATL STAGE PREDICTOR | 44 | 24 | All SZGR 2.0 genes in this pathway |
| GALLUZZI PREVENT MITOCHONDIAL PERMEABILIZATION | 22 | 16 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| MENSSEN MYC TARGETS | 53 | 35 | All SZGR 2.0 genes in this pathway |
| IIZUKA LIVER CANCER PROGRESSION L0 L1 DN | 21 | 14 | All SZGR 2.0 genes in this pathway |
| YAGI AML RELAPSE PROGNOSIS | 35 | 24 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING DN | 68 | 44 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| YAGI AML SURVIVAL | 129 | 87 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH3 21 DN | 49 | 29 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE UP | 156 | 92 | All SZGR 2.0 genes in this pathway |
| WONG MITOCHONDRIA GENE MODULE | 217 | 122 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO FLUOROURACIL | 17 | 8 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| VANLOO SP3 TARGETS DN | 89 | 47 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP | 149 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP | 289 | 184 | All SZGR 2.0 genes in this pathway |