Gene Page: THRA
Summary ?
| GeneID | 7067 |
| Symbol | THRA |
| Synonyms | AR7|CHNG6|EAR7|ERB-T-1|ERBA|ERBA1|NR1A1|THRA1|THRA2|c-ERBA-1 |
| Description | thyroid hormone receptor, alpha |
| Reference | MIM:190120|HGNC:HGNC:11796|Ensembl:ENSG00000126351|HPRD:07185|Vega:OTTHUMG00000133332 |
| Gene type | protein-coding |
| Map location | 17q11.2 |
| Pascal p-value | 0.042 |
| Sherlock p-value | 0.59 |
| Fetal beta | -0.269 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23121350 | 17 | 38219361 | THRA | 4.79E-8 | -0.014 | 1.29E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17542055 | chr7 | 42652803 | THRA | 7067 | 0.15 | trans |
Section II. Transcriptome annotation
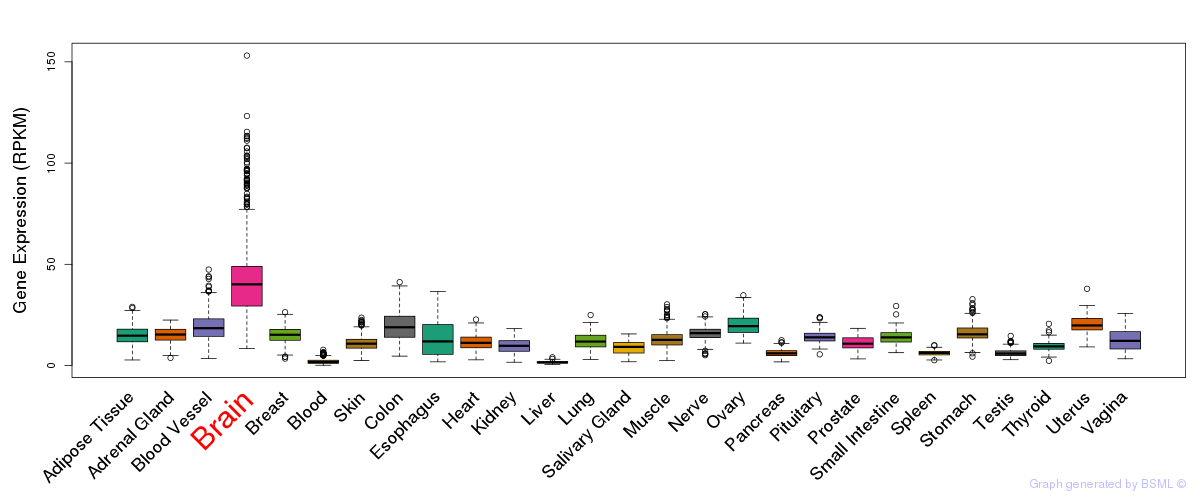
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| UTP14C | 0.96 | 0.97 |
| MTDH | 0.94 | 0.95 |
| AC012621.2 | 0.94 | 0.95 |
| TRIP12 | 0.94 | 0.95 |
| PRKAA1 | 0.94 | 0.95 |
| BCLAF1 | 0.93 | 0.95 |
| GTF2A1 | 0.93 | 0.95 |
| ZNF507 | 0.93 | 0.96 |
| BAG5 | 0.93 | 0.94 |
| CEP97 | 0.93 | 0.96 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.72 | -0.79 |
| FXYD1 | -0.71 | -0.79 |
| MT-CO2 | -0.71 | -0.80 |
| HIGD1B | -0.69 | -0.78 |
| IFI27 | -0.69 | -0.78 |
| ENHO | -0.68 | -0.80 |
| MT-CYB | -0.67 | -0.75 |
| AF347015.21 | -0.67 | -0.77 |
| AF347015.8 | -0.67 | -0.76 |
| AF347015.33 | -0.67 | -0.73 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKAP13 | AKAP-Lbc | BRX | FLJ11952 | FLJ43341 | HA-3 | Ht31 | LBC | PROTO-LB | PROTO-LBC | c-lbc | A kinase (PRKA) anchor protein 13 | - | HPRD,BioGRID | 9627117 |
| BTG1 | - | B-cell translocation gene 1, anti-proliferative | BTG1 interacts with THRA (TR-alpha-1). This interaction was modeled on a demonstrated interaction between human BTG1 and chicken or quail TR-alpha-1. | BIND | 15674337 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | - | HPRD,BioGRID | 12048199 |
| CDK7 | CAK1 | CDKN7 | MO15 | STK1 | p39MO15 | cyclin-dependent kinase 7 | TR-alpha interacts with hMo15. This interaction was modeled on a demonstrated interaction between chicken TR-alpha and human hMo15. | BIND | 15249124 |
| CDK8 | K35 | MGC126074 | MGC126075 | cyclin-dependent kinase 8 | Affinity Capture-Western | BioGRID | 10198638 |
| COPS2 | ALIEN | CSN2 | SGN2 | TRIP15 | COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis) | - | HPRD,BioGRID | 10207062 |
| CREB1 | CREB | MGC9284 | cAMP responsive element binding protein 1 | - | HPRD | 12805224 |
| DAP3 | DAP-3 | DKFZp686G12159 | MGC126058 | MGC126059 | MRP-S29 | MRPS29 | bMRP-10 | death associated protein 3 | Reconstituted Complex | BioGRID | 10903152 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | Affinity Capture-Western | BioGRID | 12371907 |
| FUS | CHOP | FUS-CHOP | FUS1 | TLS | TLS/CHOP | hnRNP-P2 | fusion (involved in t(12;16) in malignant liposarcoma) | - | HPRD,BioGRID | 9440806 |
| GTF2B | TF2B | TFIIB | general transcription factor IIB | GTF2B (TFIIB) interacts with THRA (T3R-alpha). This interaction was modelled on a demonstrated interaction between human TFIIB and chicken T3R-alpha. | BIND | 7609079 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | - | HPRD | 10508171 |
| HR | ALUNC | AU | HSA277165 | hairless homolog (mouse) | Affinity Capture-Western | BioGRID | 11641275 |
| ITGB3BP | CENP-R | CENPR | HSU37139 | NRIF3 | TAP20 | integrin beta 3 binding protein (beta3-endonexin) | Reconstituted Complex Two-hybrid | BioGRID | 10490654 |11713274 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | - | HPRD,BioGRID | 9653119 |
| MED12 | CAGH45 | HOPA | KIAA0192 | OPA1 | TNRC11 | TRAP230 | mediator complex subunit 12 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 10198638 |
| MED16 | DRIP92 | THRAP5 | TRAP95 | mediator complex subunit 16 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 10198638 |
| MED21 | SRB7 | SURB7 | mediator complex subunit 21 | TR-alpha interacts with hSrb7. This interaction was modeled on a demonstrated interaction between chicken TR-alpha and human hSrb7. | BIND | 15249124 |
| MED21 | SRB7 | SURB7 | mediator complex subunit 21 | Affinity Capture-Western | BioGRID | 10198638 |
| MED24 | ARC100 | CRSP100 | CRSP4 | DRIP100 | KIAA0130 | MGC8748 | THRAP4 | TRAP100 | mediator complex subunit 24 | Affinity Capture-Western | BioGRID | 10198638 |
| MED6 | NY-REN-28 | mediator complex subunit 6 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 10198638 |
| MEF2A | ADCAD1 | RSRFC4 | RSRFC9 | myocyte enhancer factor 2A | - | HPRD,BioGRID | 12371907 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | TR-alpha interacts with SRC-1. This interaction was modeled on a demonstrated interaction between chicken TR-alpha and human SRC-1. | BIND | 15249124 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 10567404 |10847592 |11158331 |
| NR0B2 | FLJ17090 | SHP | SHP1 | nuclear receptor subfamily 0, group B, member 2 | Reconstituted Complex | BioGRID | 8650544 |
| NR2F1 | COUP-TFI | EAR-3 | EAR3 | ERBAL3 | NR2F2 | SVP44 | TCFCOUP1 | TFCOUP1 | nuclear receptor subfamily 2, group F, member 1 | - | HPRD | 1331778 |
| NRIP1 | FLJ77253 | RIP140 | nuclear receptor interacting protein 1 | Reconstituted Complex | BioGRID | 9626662 |
| NSD1 | ARA267 | DKFZp666C163 | FLJ10684 | FLJ22263 | FLJ44628 | KMT3B | SOTOS | STO | nuclear receptor binding SET domain protein 1 | - | HPRD | 9628876 |
| PARP1 | ADPRT | ADPRT1 | PARP | PARP-1 | PPOL | pADPRT-1 | poly (ADP-ribose) polymerase 1 | PARP-1 interacts with TR-alpha. This interaction was modeled on a demonstrated interaction between human PARP-1 and TR-alpha from an unspecified species. | BIND | 15808511 |
| PCSK2 | NEC2 | PC2 | SPC2 | proprotein convertase subtilisin/kexin type 2 | THRA interacts with the PCSK2 (PC2) promoter. This interaction was modelled on a demonstrated interaction between chicken THRA and human PCSK2 (PC2) promoter. | BIND | 10965896 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD | 10610177 |
| POU2F1 | OCT1 | OTF1 | POU class 2 homeobox 1 | Reconstituted Complex | BioGRID | 10480874 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | Co-purification | BioGRID | 1314167 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | - | HPRD | 1331778 |
| RXRB | DAUDI6 | H-2RIIBP | MGC1831 | NR2B2 | RCoR-1 | retinoid X receptor, beta | - | HPRD | 1331778 |
| TDG | - | thymine-DNA glycosylase | - | HPRD,BioGRID | 12874288 |
| TRIM24 | PTC6 | RNF82 | TF1A | TIF1 | TIF1A | TIF1ALPHA | hTIF1 | tripartite motif-containing 24 | Reconstituted Complex | BioGRID | 9115274 |
| TRIP11 | CEV14 | GMAP-210 | TRIP230 | thyroid hormone receptor interactor 11 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9256431 |
| YWHAH | YWHA1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | - | HPRD | 11266503 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | Reconstituted Complex | BioGRID | 11266503 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EGFR SMRTE PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| PID RXR VDR PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | 49 | 36 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| CHIN BREAST CANCER COPY NUMBER UP | 27 | 18 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| LIU LIVER CANCER | 38 | 18 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q11 Q21 AMPLICON | 133 | 78 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| JOSEPH RESPONSE TO SODIUM BUTYRATE DN | 64 | 45 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS DN | 135 | 88 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL DN | 127 | 75 | All SZGR 2.0 genes in this pathway |
| HOFFMANN PRE BI TO LARGE PRE BII LYMPHOCYTE DN | 75 | 61 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC UP | 72 | 53 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |