Gene Page: TLE1
Summary ?
| GeneID | 7088 |
| Symbol | TLE1 |
| Synonyms | ESG|ESG1|GRG1 |
| Description | transducin like enhancer of split 1 |
| Reference | MIM:600189|HGNC:HGNC:11837|Ensembl:ENSG00000196781|HPRD:02557|Vega:OTTHUMG00000021008 |
| Gene type | protein-coding |
| Map location | 9q21.32 |
| Pascal p-value | 4.704E-4 |
| Sherlock p-value | 0.813 |
| Fetal beta | 0.211 |
| DMG | 1 (# studies) |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14095100 | 9 | 84303413 | TLE1 | 2.775E-4 | 0.665 | 0.039 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
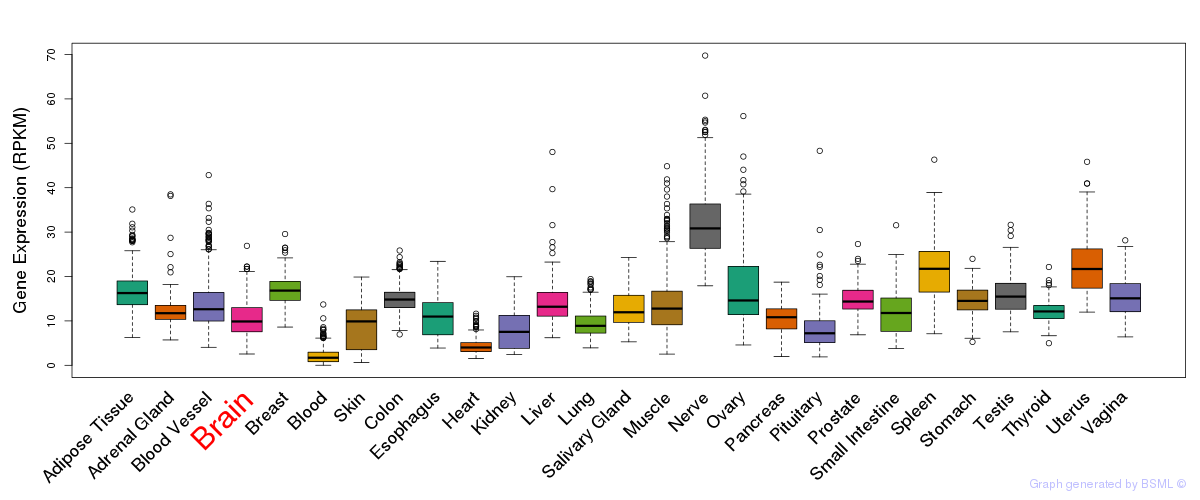
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AES | AES-1 | AES-2 | ESP1 | GRG | GRG5 | TLE5 | amino-terminal enhancer of split | Affinity Capture-MS | BioGRID | 17353931 |
| ARL3 | ARFL3 | ADP-ribosylation factor-like 3 | Two-hybrid | BioGRID | 16169070 |
| ARL4D | ARF4L | ARL6 | ADP-ribosylation factor-like 4D | Two-hybrid | BioGRID | 16169070 |
| ATN1 | B37 | D12S755E | DRPLA | NOD | atrophin 1 | Two-hybrid | BioGRID | 16169070 |
| BTBD2 | - | BTB (POZ) domain containing 2 | Two-hybrid | BioGRID | 16169070 |
| CCL18 | AMAC-1 | AMAC1 | CKb7 | DC-CK1 | DCCK1 | MIP-4 | PARC | SCYA18 | chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) | Two-hybrid | BioGRID | 16169070 |
| CCNB1 | CCNB | cyclin B1 | - | HPRD | 12397081 |
| CDC2 | CDC28A | CDK1 | DKFZp686L20222 | MGC111195 | cell division cycle 2, G1 to S and G2 to M | - | HPRD | 12397081 |
| CDKN2C | INK4C | p18 | p18-INK4C | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | Two-hybrid | BioGRID | 16169070 |
| CRCT1 | C1orf42 | NICE-1 | cysteine-rich C-terminal 1 | Two-hybrid | BioGRID | 16169070 |
| DLEU1 | BCMS | DLB1 | DLEU2 | LEU1 | LEU2 | MGC22430 | NCRNA00021 | XTP6 | deleted in lymphocytic leukemia 1 (non-protein coding) | Two-hybrid | BioGRID | 16169070 |
| EIF2S2 | DKFZp686L18198 | EIF2 | EIF2B | EIF2beta | MGC8508 | eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa | Two-hybrid | BioGRID | 16169070 |
| ERH | DROER | FLJ27340 | enhancer of rudimentary homolog (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| FOXA2 | HNF3B | MGC19807 | TCF3B | forkhead box A2 | - | HPRD,BioGRID | 10748198 |
| FOXG1 | BF1 | BF2 | FHKL3 | FKH2 | FKHL1 | FKHL2 | FKHL3 | FKHL4 | FOXG1A | FOXG1B | FOXG1C | HBF-1 | HBF-2 | HBF-3 | HBF-G2 | HBF2 | HFK1 | HFK2 | HFK3 | KHL2 | QIN | forkhead box G1 | - | HPRD | 11238932 |
| FUBP1 | FBP | FUBP | far upstream element (FUSE) binding protein 1 | Two-hybrid | BioGRID | 16169070 |
| FXYD6 | - | FXYD domain containing ion transport regulator 6 | Two-hybrid | BioGRID | 16169070 |
| GSTM4 | GSTM4-4 | GTM4 | MGC131945 | MGC9247 | glutathione S-transferase mu 4 | Two-hybrid | BioGRID | 16169070 |
| HES1 | FLJ20408 | HES-1 | HHL | HRY | bHLHb39 | hairy and enhancer of split 1, (Drosophila) | - | HPRD | 8687460 |
| HES6 | bHLHc23 | hairy and enhancer of split 6 (Drosophila) | - | HPRD,BioGRID | 11551980 |
| HHEX | HEX | HMPH | HOX11L-PEN | PRH | PRHX | hematopoietically expressed homeobox | HHEX (PRH) interacts with TLE1. | BIND | 15187083 |
| HLA-DQA1 | CD | CELIAC1 | DQ-A1 | FLJ27088 | FLJ27328 | GSE | HLA-DQA | MGC149527 | major histocompatibility complex, class II, DQ alpha 1 | Two-hybrid | BioGRID | 16169070 |
| HMGB1 | DKFZp686A04236 | HMG1 | HMG3 | SBP-1 | high-mobility group box 1 | - | HPRD,BioGRID | 11748221 |
| HNF1A | HNF1 | LFB1 | MODY3 | TCF1 | HNF1 homeobox A | - | HPRD,BioGRID | 10209158 |
| HSPE1 | CPN10 | GROES | HSP10 | heat shock 10kDa protein 1 (chaperonin 10) | Two-hybrid | BioGRID | 16169070 |
| IL6ST | CD130 | CDw130 | GP130 | GP130-RAPS | IL6R-beta | interleukin 6 signal transducer (gp130, oncostatin M receptor) | - | HPRD,BioGRID | 12030375 |
| LEF1 | DKFZp586H0919 | TCF1ALPHA | lymphoid enhancer-binding factor 1 | - | HPRD,BioGRID | 9751710 |
| LEF1 | DKFZp586H0919 | TCF1ALPHA | lymphoid enhancer-binding factor 1 | LEF-1 interacts with TLE1. This interaction was modeled on a demonstrated interaction between LEF-1 from an unspecified species and human TLE1. | BIND | 9751710 |
| MPHOSPH6 | MPP | MPP-6 | MPP6 | M-phase phosphoprotein 6 | Two-hybrid | BioGRID | 16169070 |
| PAFAH1B3 | - | platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa | Two-hybrid | BioGRID | 16169070 |
| PCDHA4 | CNR1 | CNRN1 | CRNR1 | MGC138307 | MGC142169 | PCDH-ALPHA4 | protocadherin alpha 4 | Two-hybrid | BioGRID | 16169070 |
| PFN1 | - | profilin 1 | Two-hybrid | BioGRID | 16169070 |
| POLB | MGC125976 | polymerase (DNA directed), beta | Two-hybrid | BioGRID | 16169070 |
| POLE2 | DPE2 | polymerase (DNA directed), epsilon 2 (p59 subunit) | Two-hybrid | BioGRID | 16169070 |
| PRDM1 | BLIMP1 | MGC118922 | MGC118923 | MGC118924 | MGC118925 | PRDI-BF1 | PR domain containing 1, with ZNF domain | - | HPRD,BioGRID | 9887105 |
| PSMD11 | MGC3844 | Rpn6 | S9 | p44.5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | Two-hybrid | BioGRID | 16169070 |
| PXMP3 | PAF-1 | PAF1 | PEX2 | PMP3 | PMP35 | RNF72 | peroxisomal membrane protein 3, 35kDa | Two-hybrid | BioGRID | 16169070 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | TLE1 interacts with p65. | BIND | 10660609 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | Affinity Capture-Western | BioGRID | 10660609 |
| RNF10 | KIAA0262 | MGC126758 | MGC126764 | RIE2 | ring finger protein 10 | Two-hybrid | BioGRID | 16169070 |
| RPA2 | REPA2 | RPA32 | replication protein A2, 32kDa | Two-hybrid | BioGRID | 16169070 |
| RUNX1 | AML1 | AML1-EVI-1 | AMLCR1 | CBFA2 | EVI-1 | PEBP2aB | runt-related transcription factor 1 | AML1a interacts with TLE1. | BIND | 9751710 |
| RUNX1 | AML1 | AML1-EVI-1 | AMLCR1 | CBFA2 | EVI-1 | PEBP2aB | runt-related transcription factor 1 | - | HPRD,BioGRID | 9751710 |
| RUNX1 | AML1 | AML1-EVI-1 | AMLCR1 | CBFA2 | EVI-1 | PEBP2aB | runt-related transcription factor 1 | AML1b interacts with TLE1. | BIND | 9751710 |
| RUNX3 | AML2 | CBFA3 | FLJ34510 | MGC16070 | PEBP2aC | runt-related transcription factor 3 | AML2 interacts with TLE1. | BIND | 9751710 |
| RUNX3 | AML2 | CBFA3 | FLJ34510 | MGC16070 | PEBP2aC | runt-related transcription factor 3 | - | HPRD,BioGRID | 9751710 |
| SAT1 | DC21 | KFSD | SAT | SSAT | SSAT-1 | spermidine/spermine N1-acetyltransferase 1 | Two-hybrid | BioGRID | 16169070 |
| SERPINB9 | CAP-3 | CAP3 | PI9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 | Two-hybrid | BioGRID | 16169070 |
| SIX1 | BOS3 | DFNA23 | TIP39 | SIX homeobox 1 | SIX1 interacts with TLE1. | BIND | 12441302 |
| SIX2 | - | SIX homeobox 2 | SIX2 interacts with TLE1. This interaction was modeled on a demonstrated interaction between mouse Six2 and human TLE1. | BIND | 12441302 |
| SIX3 | HPE2 | SIX homeobox 3 | SIX3 interacts with TLE1. | BIND | 12441302 |
| SIX3 | HPE2 | SIX homeobox 3 | - | HPRD,BioGRID | 12441302 |
| SIX4 | AREC3 | MGC119450 | MGC119452 | MGC119453 | SIX homeobox 4 | SIX4 interacts with TLE1. This interaction was modeled on a demonstrated interaction between mouse Six4 and human TLE1. | BIND | 12441302 |
| SIX6 | MCOPCT2 | OPTX2 | Six9 | SIX homeobox 6 | SIX6 interacts with TLE1. | BIND | 12441302 |
| SIX6 | MCOPCT2 | OPTX2 | Six9 | SIX homeobox 6 | Reconstituted Complex Two-hybrid | BioGRID | 12441302 |
| SNRPG | MGC117317 | SMG | small nuclear ribonucleoprotein polypeptide G | Two-hybrid | BioGRID | 16169070 |
| TK1 | TK2 | thymidine kinase 1, soluble | Two-hybrid | BioGRID | 16169070 |
| TLE2 | ESG | ESG2 | FLJ41188 | GRG2 | transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) | - | HPRD,BioGRID | 9874198 |
| TNRC4 | BRUNOL1 | CAGH4 | CELF3 | ERDA4 | MGC57297 | trinucleotide repeat containing 4 | Two-hybrid | BioGRID | 16169070 |
| UTX | DKFZp686A03225 | MGC141941 | bA386N14.2 | ubiquitously transcribed tetratricopeptide repeat, X chromosome | - | HPRD,BioGRID | 9854018 |
| UTY | DKFZp686L12190 | UTY1 | ubiquitously transcribed tetratricopeptide repeat gene, Y-linked | - | HPRD,BioGRID | 9854018 |
| ZFP64 | MGC940 | ZNF338 | zinc finger protein 64 homolog (mouse) | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA WNT PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| PID PS1 PATHWAY | 46 | 39 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID HES HEY PATHWAY | 48 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | 46 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH1 | 70 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS UP | 137 | 94 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO PACLITAXEL VIA MAPK8 UP | 14 | 9 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| LIU COMMON CANCER GENES | 79 | 47 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR UP | 116 | 79 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT AND CANCER BOX1 UP | 15 | 10 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT AND CANCER BOX2 DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT AND CANCER BOX5 DN | 8 | 5 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER SOX9 TARGETS IN PROSTATE DEVELOPMENT UP | 21 | 19 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH PML RARA FUSION | 77 | 62 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL CURED VS FATAL DN | 45 | 30 | All SZGR 2.0 genes in this pathway |
| WILLERT WNT SIGNALING | 24 | 13 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| OKUMURA INFLAMMATORY RESPONSE LPS | 183 | 115 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH MUTATED VH GENES | 18 | 14 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX UP | 83 | 66 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 2 | 72 | 52 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D7 | 40 | 21 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO OR UV | 63 | 44 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE DN | 88 | 42 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| GRADE METASTASIS DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| BEIER GLIOMA STEM CELL UP | 39 | 17 | All SZGR 2.0 genes in this pathway |
| SETLUR PROSTATE CANCER TMPRSS2 ERG FUSION UP | 67 | 48 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA1 DN | 74 | 47 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 DN | 88 | 61 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 DN | 82 | 51 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE MIDDLE | 98 | 56 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |