Gene Page: VHL
Summary ?
| GeneID | 7428 |
| Symbol | VHL |
| Synonyms | HRCA1|RCA1|VHL1|pVHL |
| Description | von Hippel-Lindau tumor suppressor |
| Reference | MIM:608537|HGNC:HGNC:12687|HPRD:01905| |
| Gene type | protein-coding |
| Map location | 3p25.3 |
| Pascal p-value | 0.169 |
| Sherlock p-value | 0.878 |
| Fetal beta | 1.691 |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1204 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10497672 | 0 | VHL | 7428 | 0.17 | trans | |||
| rs11230726 | chr11 | 61275103 | VHL | 7428 | 0.13 | trans |
Section II. Transcriptome annotation
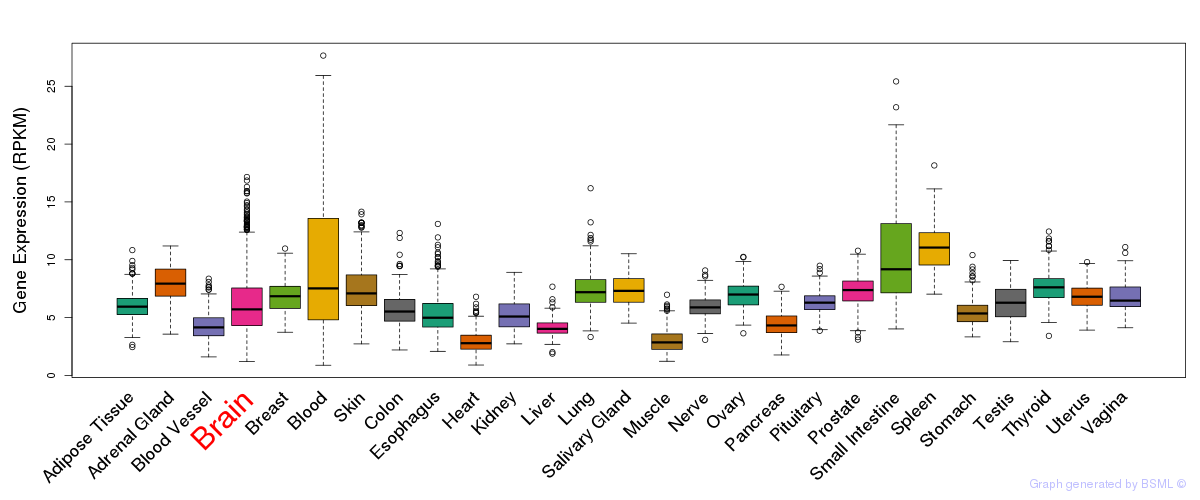
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC009041.1 | 0.70 | 0.53 |
| TRIM17 | 0.69 | 0.79 |
| AC009041.3 | 0.66 | 0.50 |
| CORT | 0.65 | 0.73 |
| NPAS2 | 0.65 | 0.73 |
| TTC22 | 0.65 | 0.69 |
| TNK1 | 0.64 | 0.24 |
| ANKRD55 | 0.63 | 0.77 |
| FHOD1 | 0.63 | 0.54 |
| APITD1 | 0.63 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HEBP2 | -0.36 | -0.61 |
| CSRP2 | -0.32 | -0.48 |
| METRNL | -0.31 | -0.47 |
| COTL1 | -0.31 | -0.25 |
| SNX7 | -0.31 | -0.26 |
| RNF182 | -0.31 | -0.37 |
| CBS | -0.30 | -0.32 |
| NEUROD6 | -0.30 | -0.17 |
| SLA | -0.30 | -0.11 |
| KLHL1 | -0.30 | -0.10 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008134 | transcription factor binding | TAS | 7660122 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | TAS | 7660122 | |
| GO:0000902 | cell morphogenesis | NAS | 12169691 | |
| GO:0001525 | angiogenesis | IEA | - | |
| GO:0006508 | proteolysis | TAS | 10353251 | |
| GO:0006511 | ubiquitin-dependent protein catabolic process | IEA | - | |
| GO:0006950 | response to stress | NAS | 12169691 | |
| GO:0008285 | negative regulation of cell proliferation | TAS | 7660130 | |
| GO:0006916 | anti-apoptosis | NAS | 12169691 | |
| GO:0016567 | protein ubiquitination | IMP | 12169691 | |
| GO:0050821 | protein stabilization | NAS | 12169691 | |
| GO:0030163 | protein catabolic process | IEA | - | |
| GO:0030198 | extracellular matrix organization | IEA | - | |
| GO:0045786 | negative regulation of cell cycle | IEA | - | |
| GO:0043534 | blood vessel endothelial cell migration | IEA | - | |
| GO:0045597 | positive regulation of cell differentiation | NAS | 12169691 | |
| GO:0045449 | regulation of transcription | IMP | 15824735 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | TAS | 7604013 | |
| GO:0005634 | nucleus | TAS | 7604013 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005739 | mitochondrion | NAS | 12169691 | |
| GO:0005783 | endoplasmic reticulum | NAS | 12169691 | |
| GO:0016020 | membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APRT | AMP | DKFZp686D13177 | MGC125856 | MGC125857 | MGC129961 | adenine phosphoribosyltransferase | Affinity Capture-MS | BioGRID | 17353931 |
| ATXN2 | ATX2 | FLJ46772 | SCA2 | TNRC13 | ataxin 2 | Affinity Capture-MS | BioGRID | 17353931 |
| CAB39 | CGI-66 | FLJ22682 | MO25 | calcium binding protein 39 | Affinity Capture-MS | BioGRID | 17353931 |
| CCDC59 | BR22 | DKFZp686K1021 | FLJ10294 | HSPC128 | TAP26 | coiled-coil domain containing 59 | Affinity Capture-MS | BioGRID | 17353931 |
| CCDC82 | FLJ23518 | HSPC048 | coiled-coil domain containing 82 | Affinity Capture-MS | BioGRID | 17353931 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | Affinity Capture-MS | BioGRID | 17353931 |
| CDO1 | - | cysteine dioxygenase, type I | Affinity Capture-MS | BioGRID | 17353931 |
| CHMP2B | CHMP2.5 | DKFZp564O123 | DMT1 | VPS2-2 | VPS2B | chromatin modifying protein 2B | Affinity Capture-MS | BioGRID | 17353931 |
| CNTF | HCNTF | ciliary neurotrophic factor | Affinity Capture-MS | BioGRID | 17353931 |
| CSTB | CST6 | EPM1 | PME | STFB | cystatin B (stefin B) | Affinity Capture-MS | BioGRID | 17353931 |
| CSTF3 | CSTF-77 | MGC117398 | MGC43001 | MGC75122 | cleavage stimulation factor, 3' pre-RNA, subunit 3, 77kDa | Affinity Capture-MS | BioGRID | 17353931 |
| CUL2 | MGC131970 | cullin 2 | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 10587522 |11384984 |17353931 |
| CUL5 | VACM-1 | VACM1 | cullin 5 | Reconstituted Complex | BioGRID | 11384984 |
| EPAS1 | ECYT4 | HIF2A | HLF | MOP2 | PASD2 | endothelial PAS domain protein 1 | The von Hippel-Lindau tumor suppressor protein (pVHL) interacts with Hypoxia inducible factor-2-alpha (HIF-2 alpha) | BIND | 10823831 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | The von Hippel-Lindau tumor suppressor protein (pVHL) interacts with FLNA . | BIND | 12169691 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | - | HPRD,BioGRID | 8674032 |12169691 |
| FN1 | CIG | DKFZp686F10164 | DKFZp686H0342 | DKFZp686I1370 | DKFZp686O13149 | ED-B | FINC | FN | FNZ | GFND | GFND2 | LETS | MSF | fibronectin 1 | pVHL interacts with fibronectin 1 | BIND | 10823831 |
| FN1 | CIG | DKFZp686F10164 | DKFZp686H0342 | DKFZp686I1370 | DKFZp686O13149 | ED-B | FINC | FN | FNZ | GFND | GFND2 | LETS | MSF | fibronectin 1 | - | HPRD,BioGRID | 9651579 |
| FSCN1 | FLJ38511 | SNL | p55 | fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) | Affinity Capture-MS | BioGRID | 17353931 |
| HAS1 | HAS | hyaluronan synthase 1 | Affinity Capture-MS | BioGRID | 17353931 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | Reconstituted Complex | BioGRID | 11641274 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Reconstituted Complex | BioGRID | 11641274 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | Reconstituted Complex | BioGRID | 11641274 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | Hydroxylated HIF-1 alpha interacts with VHL. This interaction was modeled on a demonstrated interaction between human hydroxylated HIF-1 alpha and an unspecified isoform of VHL from an unspecified species. | BIND | 15721254 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | The von Hippel-Lindau tumor suppressor protein (pVHL) interacts with Hypoxia inducible factor-alpha (HIF-1 alpha) | BIND | 10823831 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | - | HPRD | 11504942 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | HIF1A (HIF-1alpha) interacts with VHL. | BIND | 15750626 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | - | HPRD,BioGRID | 10944113 |
| HIF1AN | DKFZp762F1811 | FIH1 | FLJ20615 | FLJ22027 | hypoxia-inducible factor 1, alpha subunit inhibitor | Reconstituted Complex | BioGRID | 11641274 |
| HIF3A | HIF-3A | HIF-3A2 | HIF-3A4 | IPAS | MOP7 | PASD7 | bHLHe17 | hypoxia inducible factor 3, alpha subunit | - | HPRD | 12538644 |
| HIST1H2BC | H2B.1 | H2B/l | H2BFL | MGC104246 | dJ221C16.3 | histone cluster 1, H2bc | Affinity Capture-MS | BioGRID | 17353931 |
| HNRNPA2B1 | DKFZp779B0244 | FLJ22720 | HNRNPA2 | HNRNPB1 | HNRPA2 | HNRPA2B1 | HNRPB1 | RNPA2 | SNRPB1 | heterogeneous nuclear ribonucleoprotein A2/B1 | - | HPRD,BioGRID | 11517223 |
| IARS | FLJ20736 | IARS1 | ILRS | PRO0785 | isoleucyl-tRNA synthetase | Affinity Capture-MS | BioGRID | 17353931 |
| IMPDH2 | IMPD2 | IMPDH-II | IMP (inosine monophosphate) dehydrogenase 2 | Affinity Capture-MS | BioGRID | 17353931 |
| JMJD1C | DKFZp761F0118 | FLJ14374 | KIAA1380 | RP11-10C13.2 | TRIP8 | jumonji domain containing 1C | Affinity Capture-MS | BioGRID | 17353931 |
| KNTC1 | FLJ36151 | KIAA0166 | ROD | kinetochore associated 1 | Affinity Capture-MS | BioGRID | 17353931 |
| MCC | DKFZp762O1615 | FLJ38893 | FLJ46755 | MCC1 | mutated in colorectal cancers | Affinity Capture-MS | BioGRID | 17353931 |
| MOBKL3 | 2C4D | CGI-95 | MGC12264 | MOB1 | MOB3 | PREI3 | MOB1, Mps One Binder kinase activator-like 3 (yeast) | Affinity Capture-MS | BioGRID | 17353931 |
| MRCL3 | MLCB | MRLC3 | myosin regulatory light chain MRCL3 | Affinity Capture-MS | BioGRID | 17353931 |
| NEDD8 | FLJ43224 | MGC104393 | MGC125896 | MGC125897 | Nedd-8 | neural precursor cell expressed, developmentally down-regulated 8 | Affinity Capture-MS | BioGRID | 17353931 |
| PAPSS2 | ATPSK2 | SK2 | 3'-phosphoadenosine 5'-phosphosulfate synthase 2 | Affinity Capture-MS | BioGRID | 17353931 |
| PCMT1 | - | protein-L-isoaspartate (D-aspartate) O-methyltransferase | Affinity Capture-MS | BioGRID | 17353931 |
| PFAS | FGAMS | FGARAT | KIAA0361 | PURL | phosphoribosylformylglycinamidine synthase | Affinity Capture-MS | BioGRID | 17353931 |
| PHF17 | FLJ22479 | JADE1 | KIAA1807 | PHD finger protein 17 | The von Hippel-Lindau tumor suppressor protein (pVHL) interacts with Jade-1 | BIND | 12169691 |
| PHF17 | FLJ22479 | JADE1 | KIAA1807 | PHD finger protein 17 | - | HPRD,BioGRID | 12169691 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | Affinity Capture-MS | BioGRID | 17353931 |
| PKD1L3 | - | polycystic kidney disease 1-like 3 | Affinity Capture-MS | BioGRID | 17353931 |
| PPIB | CYP-S1 | CYPB | MGC14109 | MGC2224 | SCYLP | peptidylprolyl isomerase B (cyclophilin B) | Affinity Capture-MS | BioGRID | 17353931 |
| PSMB1 | FLJ25321 | HC5 | KIAA1838 | PMSB1 | PSC5 | proteasome (prosome, macropain) subunit, beta type, 1 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMB3 | HC10-II | MGC4147 | proteasome (prosome, macropain) subunit, beta type, 3 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMC3 | MGC8487 | TBP1 | proteasome (prosome, macropain) 26S subunit, ATPase, 3 | - | HPRD,BioGRID | 14556007 |
| PSMD13 | HSPC027 | Rpn9 | S11 | p40.5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 | Affinity Capture-MS | BioGRID | 17353931 |
| RAB1B | - | RAB1B, member RAS oncogene family | Affinity Capture-MS | BioGRID | 17353931 |
| RASGRP1 | CALDAG-GEFI | CALDAG-GEFII | MGC129998 | MGC129999 | RASGRP | V | hRasGRP1 | RAS guanyl releasing protein 1 (calcium and DAG-regulated) | Affinity Capture-MS | BioGRID | 17353931 |
| RBX1 | BA554C12.1 | MGC13357 | MGC1481 | RNF75 | ROC1 | ring-box 1 | Affinity Capture-MS | BioGRID | 17353931 |
| RHOC | ARH9 | ARHC | H9 | MGC1448 | MGC61427 | RHOH9 | ras homolog gene family, member C | Affinity Capture-MS | BioGRID | 17353931 |
| RNF139 | HRCA1 | MGC31961 | RCA1 | TRC8 | ring finger protein 139 | - | HPRD | 12032852 |
| RPA3 | REPA3 | replication protein A3, 14kDa | Affinity Capture-MS | BioGRID | 17353931 |
| RPS9 | - | ribosomal protein S9 | Affinity Capture-MS | BioGRID | 17353931 |
| STK16 | FLJ39635 | KRCT | MPSK | PKL12 | TSF1 | serine/threonine kinase 16 | Affinity Capture-MS | BioGRID | 17353931 |
| TAGLN2 | HA1756 | KIAA0120 | transgelin 2 | Affinity Capture-MS | BioGRID | 17353931 |
| TARS | MGC9344 | ThrRS | threonyl-tRNA synthetase | Affinity Capture-MS | BioGRID | 17353931 |
| TCEB1 | SIII | transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) | Affinity Capture-MS Co-crystal Structure Reconstituted Complex Two-hybrid | BioGRID | 8674032 |10587522 |11739384 |12004076 |17353931 |
| TCEB1 | SIII | transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) | - | HPRD | 7660130 |10449727 |12004076 |
| TCEB2 | ELOB | SIII | transcription elongation factor B (SIII), polypeptide 2 (18kDa, elongin B) | Affinity Capture-MS Affinity Capture-Western Co-crystal Structure Reconstituted Complex | BioGRID | 10587522 |12004076 |17353931 |
| TGFB1I1 | ARA55 | HIC-5 | HIC5 | TSC-5 | transforming growth factor beta 1 induced transcript 1 | Affinity Capture-MS | BioGRID | 17353931 |
| TPT1 | FLJ27337 | HRF | TCTP | p02 | tumor protein, translationally-controlled 1 | Affinity Capture-MS | BioGRID | 17353931 |
| TRIM28 | FLJ29029 | KAP1 | RNF96 | TF1B | TIF1B | tripartite motif-containing 28 | Reconstituted Complex | BioGRID | 12682018 |
| TXN | DKFZp686B1993 | MGC61975 | TRX | TRX1 | thioredoxin | Affinity Capture-MS | BioGRID | 17353931 |
| USP33 | KIAA1097 | MGC16868 | VDU1 | ubiquitin specific peptidase 33 | pVHL interacts with VDU1-type II. | BIND | 11739384 |
| USP33 | KIAA1097 | MGC16868 | VDU1 | ubiquitin specific peptidase 33 | - | HPRD,BioGRID | 11739384 |
| VBP1 | PFD3 | PFDN3 | VBP-1 | von Hippel-Lindau binding protein 1 | Affinity Capture-Western Two-hybrid | BioGRID | 8674032 |12169691 |
| VBP1 | PFD3 | PFDN3 | VBP-1 | von Hippel-Lindau binding protein 1 | The von Hippel-Lindau tumor suppressor protein (pVHL) interacts with VBP1. | BIND | 12169691 |
| ZNF197 | D3S1363E | P18 | VHLaK | ZKSCAN9 | ZNF166 | ZNF20 | zinc finger protein 197 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12682018 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG UBIQUITIN MEDIATED PROTEOLYSIS | 138 | 98 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG RENAL CELL CARCINOMA | 70 | 60 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIF PATHWAY | 15 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VEGF PATHWAY | 29 | 18 | All SZGR 2.0 genes in this pathway |
| PID HIF2PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID HIF1A PATHWAY | 19 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | 18 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | 212 | 129 | All SZGR 2.0 genes in this pathway |
| NOJIMA SFRP2 TARGETS UP | 31 | 23 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW DN | 165 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH UP | 147 | 101 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA UP | 146 | 104 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 2NM UP | 81 | 57 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH VS LOW UP | 6 | 6 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN | 110 | 78 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A UP | 142 | 104 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| DORSAM HOXA9 TARGETS DN | 32 | 22 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS DN | 31 | 25 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA | 140 | 96 | All SZGR 2.0 genes in this pathway |