Gene Page: YWHAH
Summary ?
| GeneID | 7533 |
| Symbol | YWHAH |
| Synonyms | YWHA1 |
| Description | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein eta |
| Reference | MIM:113508|HGNC:HGNC:12853|Ensembl:ENSG00000128245|HPRD:00215|Vega:OTTHUMG00000030833 |
| Gene type | protein-coding |
| Map location | 22q12.3 |
| Pascal p-value | 0.307 |
| Sherlock p-value | 0.709 |
| Fetal beta | -0.683 |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_mGluR5 G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.989 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
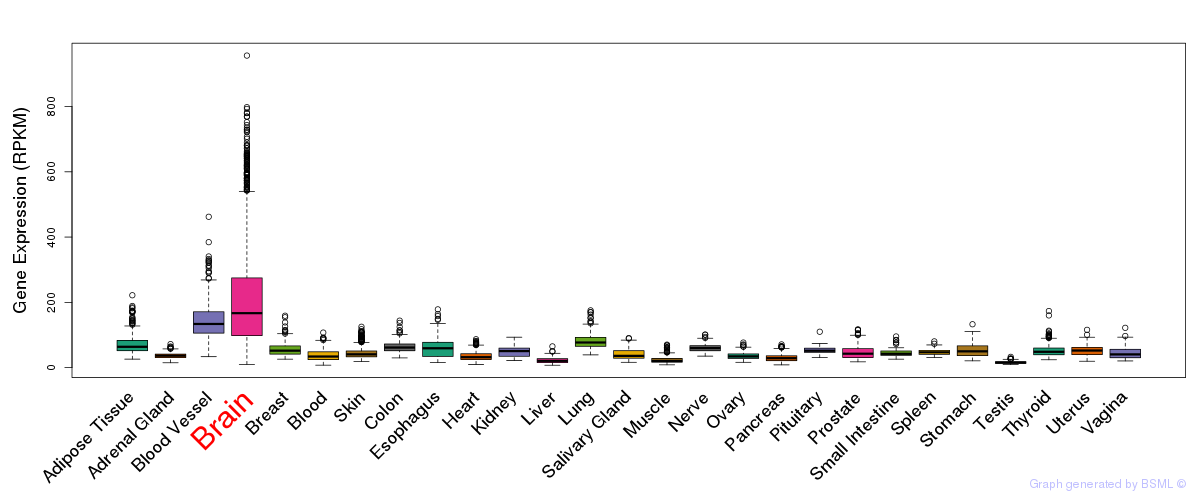
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF845 | 0.96 | 0.90 |
| SMARCAD1 | 0.95 | 0.92 |
| ZNF670 | 0.95 | 0.90 |
| ZNF468 | 0.95 | 0.91 |
| TRIM13 | 0.94 | 0.83 |
| ZNF555 | 0.94 | 0.86 |
| CCDC50 | 0.94 | 0.87 |
| PHF16 | 0.94 | 0.88 |
| GNA13 | 0.94 | 0.89 |
| TNPO1 | 0.94 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.67 | -0.74 |
| HLA-F | -0.66 | -0.75 |
| FBXO2 | -0.64 | -0.66 |
| AIFM3 | -0.63 | -0.71 |
| PTH1R | -0.63 | -0.70 |
| AC018755.7 | -0.63 | -0.70 |
| SLC16A11 | -0.63 | -0.64 |
| LHPP | -0.63 | -0.58 |
| AF347015.27 | -0.63 | -0.81 |
| TNFSF12 | -0.62 | -0.62 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003779 | actin binding | IEA | - | |
| GO:0003779 | actin binding | ISS | - | |
| GO:0005159 | insulin-like growth factor receptor binding | ISS | - | |
| GO:0019904 | protein domain specific binding | ISS | - | |
| GO:0016563 | transcription activator activity | IDA | 15790729 | |
| GO:0035259 | glucocorticoid receptor binding | IPI | 9079630 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0048167 | regulation of synaptic plasticity | ISS | Synap (GO term level: 8) | - |
| GO:0050774 | negative regulation of dendrite morphogenesis | ISS | neurogenesis, dendrite (GO term level: 14) | - |
| GO:0045664 | regulation of neuron differentiation | ISS | neuron, neurogenesis (GO term level: 9) | - |
| GO:0006713 | glucocorticoid catabolic process | IDA | 15790729 | |
| GO:0006886 | intracellular protein transport | ISS | - | |
| GO:0042921 | glucocorticoid receptor signaling pathway | IDA | 15790729 | |
| GO:0045941 | positive regulation of transcription | IDA | 15790729 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | ISS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | c-Abl interacts with 14-3-3-eta. | BIND | 15696159 |
| ARAF | A-RAF | ARAF1 | PKS2 | RAFA1 | v-raf murine sarcoma 3611 viral oncogene homolog | Affinity Capture-Western | BioGRID | 11969417 |
| BAD | BBC2 | BCL2L8 | BCL2-associated agonist of cell death | - | HPRD,BioGRID | 9369453 |
| BRAF | B-RAF1 | BRAF1 | FLJ95109 | MGC126806 | MGC138284 | RAFB1 | v-raf murine sarcoma viral oncogene homolog B1 | - | HPRD | 8668348 |
| CDC25B | - | cell division cycle 25 homolog B (S. pombe) | - | HPRD,BioGRID | 10713667 |
| CDC25C | CDC25 | cell division cycle 25 homolog C (S. pombe) | - | HPRD | 9278512 |10330186 |
| CDC25C | CDC25 | cell division cycle 25 homolog C (S. pombe) | Affinity Capture-Western | BioGRID | 11969417 |
| CDKN1B | CDKN4 | KIP1 | MEN1B | MEN4 | P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | - | HPRD | 12042314 |
| CHRNA4 | BFNC | EBN | EBN1 | FLJ95812 | NACHR | NACHRA4 | NACRA4 | cholinergic receptor, nicotinic, alpha 4 | - | HPRD,BioGRID | 11352901 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD | 11266503 |
| EPB41L3 | 4.1B | DAL-1 | DAL1 | FLJ37633 | KIAA0987 | erythrocyte membrane protein band 4.1-like 3 | - | HPRD,BioGRID | 11996670 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD | 11266503 |
| ESR2 | ER-BETA | ESR-BETA | ESRB | ESTRB | Erb | NR3A2 | estrogen receptor 2 (ER beta) | - | HPRD | 11266503 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | Affinity Capture-Western | BioGRID | 11969417 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | JNK interacts with 14-3-3-eta. | BIND | 15696159 |
| KCNH2 | ERG1 | HERG | HERG1 | Kv11.1 | LQT2 | SQT1 | potassium voltage-gated channel, subfamily H (eag-related), member 2 | - | HPRD | 11953308 |
| KIF5B | KINH | KNS | KNS1 | UKHC | kinesin family member 5B | Affinity Capture-MS Far Western | BioGRID | 11969417 |
| KLC3 | KLC2 | KLC2L | KLCt | KNS2B | kinesin light chain 3 | - | HPRD,BioGRID | 11969417 |
| KRT18 | CYK18 | K18 | keratin 18 | - | HPRD | 9524113 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | - | HPRD | 11266503 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD | 11266503 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | - | HPRD | 11266503 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | - | HPRD | 11266503 |
| NEK1 | DKFZp686D06121 | DKFZp686K12169 | KIAA1901 | MGC138800 | NY-REN-55 | NIMA (never in mitosis gene a)-related kinase 1 | NEK1 interacts with 14-3-3. | BIND | 14690447 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD,BioGRID | 9079630 |11266503 |12730237 |
| NRIP1 | FLJ77253 | RIP140 | nuclear receptor interacting protein 1 | - | HPRD | 11266503 |
| PCTK1 | FLJ16665 | PCTAIRE | PCTAIRE1 | PCTGAIRE | PCTAIRE protein kinase 1 | - | HPRD | 9799109 |
| PDK1 | - | pyruvate dehydrogenase kinase, isozyme 1 | - | HPRD | 12177059 |
| PDPK1 | MGC20087 | MGC35290 | PDK1 | PRO0461 | 3-phosphoinositide dependent protein kinase-1 | - | HPRD,BioGRID | 12177059 |
| PIAS2 | MGC102682 | MIZ1 | PIASX | PIASX-ALPHA | PIASX-BETA | SIZ2 | ZMIZ4 | miz | protein inhibitor of activated STAT, 2 | Miz1 interacts with 14-3-3 eta. | BIND | 15580267 |
| PIK3CA | MGC142161 | MGC142163 | PI3K | p110-alpha | phosphoinositide-3-kinase, catalytic, alpha polypeptide | Affinity Capture-Western | BioGRID | 11969417 |
| PRKCZ | PKC-ZETA | PKC2 | protein kinase C, zeta | Affinity Capture-Western | BioGRID | 10620507 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD,BioGRID | 8702721 |
| REM1 | GD:REM | GES | MGC48669 | REM | RAS (RAD and GEM)-like GTP-binding 1 | - | HPRD,BioGRID | 10441394 |
| RGNEF | DKFZp686P12164 | FLJ21817 | KIAA1998 | Rho-guanine nucleotide exchange factor | - | HPRD | 11533041 |
| RIMS1 | CORD7 | KIAA0340 | MGC167823 | MGC176677 | RAB3IP2 | RIM | RIM1 | regulating synaptic membrane exocytosis 1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12871946 |
| RIMS2 | DKFZp781A0653 | KIAA0751 | OBOE | RAB3IP3 | RIM2 | regulating synaptic membrane exocytosis 2 | - | HPRD,BioGRID | 12871946 |
| RPH3A | KIAA0985 | rabphilin 3A homolog (mouse) | - | HPRD,BioGRID | 12871946 |
| SNRPE | B-raf | SME | small nuclear ribonucleoprotein polypeptide E | Affinity Capture-Western | BioGRID | 11969417 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD | 8702721 |
| THRA | AR7 | EAR7 | ERB-T-1 | ERBA | ERBA1 | MGC000261 | MGC43240 | NR1A1 | THRA1 | THRA2 | c-ERBA-1 | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | - | HPRD | 11266503 |
| TLX2 | Enx | HOX11L1 | NCX | T-cell leukemia homeobox 2 | - | HPRD,BioGRID | 9738002 |
| TNFAIP3 | A20 | MGC104522 | MGC138687 | MGC138688 | OTUD7C | TNFA1P2 | tumor necrosis factor, alpha-induced protein 3 | - | HPRD,BioGRID | 8702721 |9299557 |
| TPH1 | MGC119994 | TPH | TPRH | tryptophan hydroxylase 1 | - | HPRD | 7499362 |
| TSC1 | KIAA0243 | LAM | MGC86987 | TSC | tuberous sclerosis 1 | Hamartin interacts with 14-3-3-eta. This interaction is modeled on demonstrated interaction between human hamartin and mouse 14-3-3-eta isoform. | BIND | 12176984 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | - | HPRD | 12438239 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | The tumor suppressor protein tuberin interacts with 14-3-3-eta isoform. This interaction is modeled on demonstrated interaction between human tuberin and mouse 14-3-3-eta isoform. | BIND | 12176984 |
| ZFP36 | G0S24 | GOS24 | NUP475 | RNF162A | TIS11 | TTP | zinc finger protein 36, C3H type, homolog (mouse) | - | HPRD,BioGRID | 11886850 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AKT PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G2 PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HDAC PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1R PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RB PATHWAY | 13 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHREBP2 PATHWAY | 42 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BAD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PGC1A PATHWAY | 26 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ACH PATHWAY | 16 | 13 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | 51 | 41 | All SZGR 2.0 genes in this pathway |
| ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID FOXO PATHWAY | 49 | 43 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID P38 MK2 PATHWAY | 21 | 19 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID A6B1 A6B4 INTEGRIN PATHWAY | 46 | 35 | All SZGR 2.0 genes in this pathway |
| PID INSULIN GLUCOSE PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI AKT PATHWAY | 35 | 30 | All SZGR 2.0 genes in this pathway |
| PID PI3K PLC TRK PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| DEN INTERACT WITH LCA5 | 26 | 21 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| GNATENKO PLATELET SIGNATURE | 48 | 28 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN | 187 | 122 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS UP | 69 | 41 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS UP | 206 | 118 | All SZGR 2.0 genes in this pathway |
| MOREIRA RESPONSE TO TSA UP | 28 | 23 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C0 | 107 | 72 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND TBH | 60 | 42 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN WITHOUT MGMT 48HR DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| GHO ATF5 TARGETS DN | 16 | 10 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION UP | 180 | 114 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL DN | 175 | 103 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS DN | 142 | 79 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 M | 216 | 124 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 792 | 798 | m8 | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-129-5p | 397 | 403 | m8 | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-142-5p | 493 | 499 | 1A | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-15/16/195/424/497 | 793 | 799 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-219 | 67 | 73 | 1A | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-31 | 61 | 67 | 1A | hsa-miR-31 | AGGCAAGAUGCUGGCAUAGCUG |
| miR-33 | 582 | 588 | m8 | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| hsa-miR-33 | GUGCAUUGUAGUUGCAUUG | ||||
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-338 | 172 | 178 | 1A | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| miR-365 | 166 | 172 | m8 | hsa-miR-365 | UAAUGCCCCUAAAAAUCCUUAU |
| miR-450 | 397 | 403 | 1A | hsa-miR-450 | UUUUUGCGAUGUGUUCCUAAUA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.